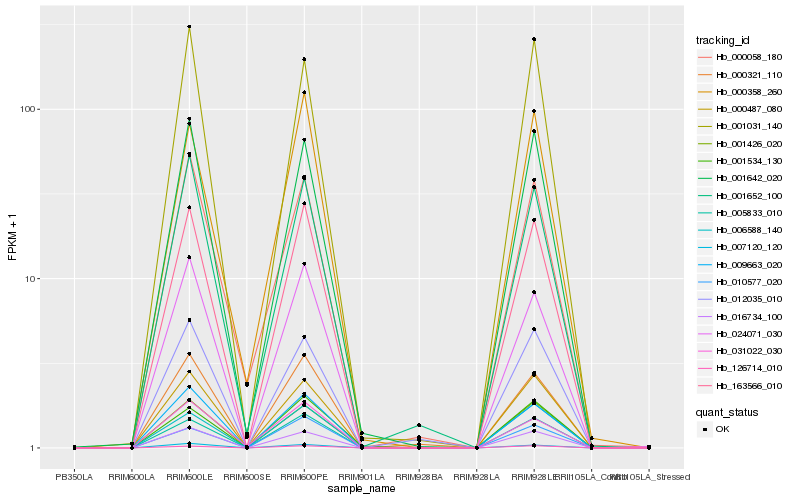
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163566_010 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 2 |
Hb_005833_010 |
0.0393871822 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 3 |
Hb_009663_020 |
0.0520014485 |
- |
- |
- |
| 4 |
Hb_016734_100 |
0.0574414762 |
- |
- |
- |
| 5 |
Hb_001426_020 |
0.0581310322 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 6 |
Hb_024071_030 |
0.0630808178 |
- |
- |
hypothetical protein CICLE_v10025479mg [Citrus clementina] |
| 7 |
Hb_000487_080 |
0.0649645405 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 8 |
Hb_007120_120 |
0.0669543537 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000321_110 |
0.0690496232 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 10 |
Hb_126714_010 |
0.0726965059 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 11 |
Hb_001642_020 |
0.076845677 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 12 |
Hb_006588_140 |
0.0791205449 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_010577_020 |
0.0843343258 |
- |
- |
- |
| 14 |
Hb_001652_100 |
0.0855683258 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 15 |
Hb_000358_260 |
0.0887528621 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 16 |
Hb_031022_030 |
0.0967789609 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 17 |
Hb_001534_130 |
0.0977590952 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 18 |
Hb_012035_010 |
0.0993156965 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 19 |
Hb_001031_140 |
0.1003904259 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 20 |
Hb_000058_180 |
0.1023895778 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |