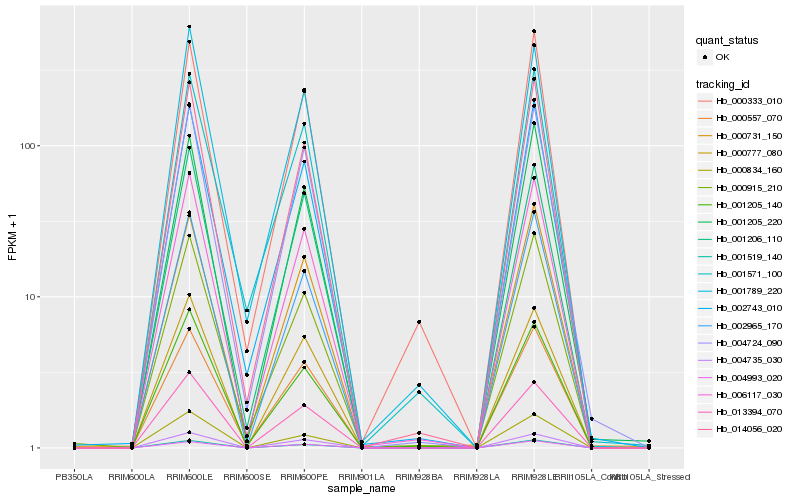
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006117_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_014056_020 |
0.0326076743 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_002965_170 |
0.0374598722 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 4 |
Hb_001205_140 |
0.041013785 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 5 |
Hb_000557_070 |
0.0411581367 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 6 |
Hb_000915_210 |
0.0434129358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004724_090 |
0.0438240136 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 8 |
Hb_001519_140 |
0.043992021 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 9 |
Hb_002743_010 |
0.0465931699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_013394_070 |
0.0470060169 |
- |
- |
hypothetical protein POPTR_0013s12090g [Populus trichocarpa] |
| 11 |
Hb_001789_220 |
0.0483588963 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 12 |
Hb_004735_030 |
0.0498538068 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 13 |
Hb_000777_080 |
0.0563518041 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 14 |
Hb_000333_010 |
0.0563909406 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 15 |
Hb_000731_150 |
0.0591332263 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 16 |
Hb_001571_100 |
0.0594623578 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 17 |
Hb_001206_110 |
0.0602554078 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 18 |
Hb_004993_020 |
0.0624414552 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_000834_160 |
0.0626311532 |
- |
- |
hypothetical protein JCGZ_07239 [Jatropha curcas] |
| 20 |
Hb_001205_220 |
0.062691155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |