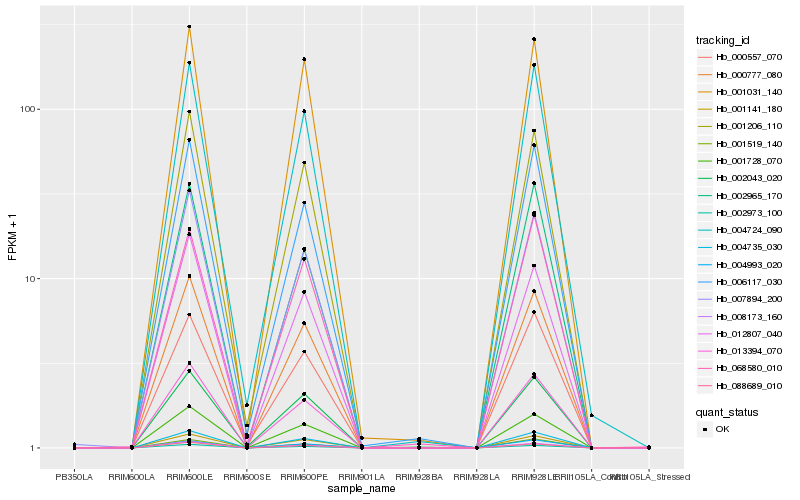
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004735_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 2 |
Hb_000777_080 |
0.0251340656 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 3 |
Hb_001141_180 |
0.0260849436 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 4 |
Hb_013394_070 |
0.0327998103 |
- |
- |
hypothetical protein POPTR_0013s12090g [Populus trichocarpa] |
| 5 |
Hb_001728_070 |
0.0336790239 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 6 |
Hb_004724_090 |
0.0415417606 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 7 |
Hb_001206_110 |
0.0429492516 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 8 |
Hb_004993_020 |
0.0461801632 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_007894_200 |
0.0472822245 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |
| 10 |
Hb_000557_070 |
0.0478643878 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 11 |
Hb_001519_140 |
0.0488302383 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 12 |
Hb_006117_030 |
0.0498538068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001031_140 |
0.051056054 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 14 |
Hb_002973_100 |
0.0513481752 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Populus euphratica] |
| 15 |
Hb_008173_160 |
0.0516964545 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 16 |
Hb_088689_010 |
0.0517054259 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 17 |
Hb_002965_170 |
0.057013245 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 18 |
Hb_002043_020 |
0.0609176543 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 19 |
Hb_068580_010 |
0.0670580361 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 20 |
Hb_012807_040 |
0.0678971221 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |