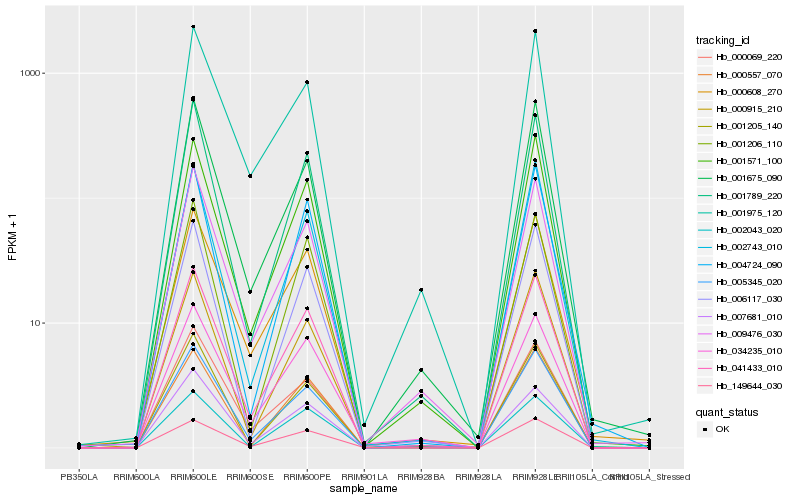
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041433_010 |
0.0 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_005345_020 |
0.0425068718 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 3 |
Hb_001789_220 |
0.0472083821 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 4 |
Hb_001571_100 |
0.0480202789 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 5 |
Hb_009476_030 |
0.0535029873 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 6 |
Hb_000608_270 |
0.0568129171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007681_010 |
0.0599490467 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 8 |
Hb_000557_070 |
0.0604364192 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 9 |
Hb_006117_030 |
0.063328613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_034235_010 |
0.0642924279 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_002043_020 |
0.0652363336 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 12 |
Hb_149644_030 |
0.0664414608 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 13 |
Hb_001206_110 |
0.0669588493 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 14 |
Hb_004724_090 |
0.0670395812 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 15 |
Hb_002743_010 |
0.0674183446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001205_140 |
0.0684612479 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 17 |
Hb_001675_090 |
0.0692434154 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001975_120 |
0.0696521633 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 19 |
Hb_000069_220 |
0.0704425208 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 20 |
Hb_000915_210 |
0.0717518649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |