| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
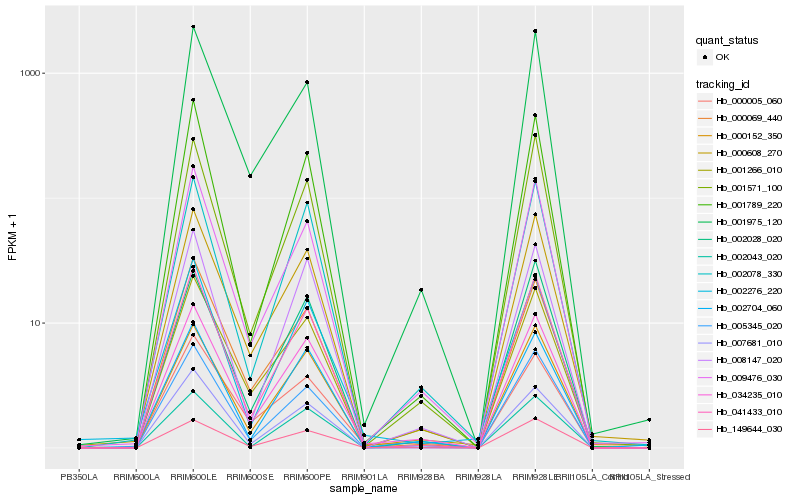
Hb_034235_010 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_149644_030 |
0.0553363761 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 3 |
Hb_001266_010 |
0.0618971471 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 4 |
Hb_041433_010 |
0.0642924279 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_009476_030 |
0.0650482643 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 6 |
Hb_002704_060 |
0.0654623563 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 7 |
Hb_000152_350 |
0.0665596217 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 8 |
Hb_002078_330 |
0.0667926993 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000608_270 |
0.067182751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005345_020 |
0.0761370707 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 11 |
Hb_000005_060 |
0.0771018447 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 12 |
Hb_008147_020 |
0.0777848181 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 13 |
Hb_002043_020 |
0.0801388032 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 14 |
Hb_001975_120 |
0.081953574 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 15 |
Hb_001571_100 |
0.0828364017 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 16 |
Hb_000069_440 |
0.0831099711 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 17 |
Hb_002276_220 |
0.0841195733 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001789_220 |
0.0873043407 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 19 |
Hb_002028_020 |
0.0889173604 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 20 |
Hb_007681_010 |
0.0890586778 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |