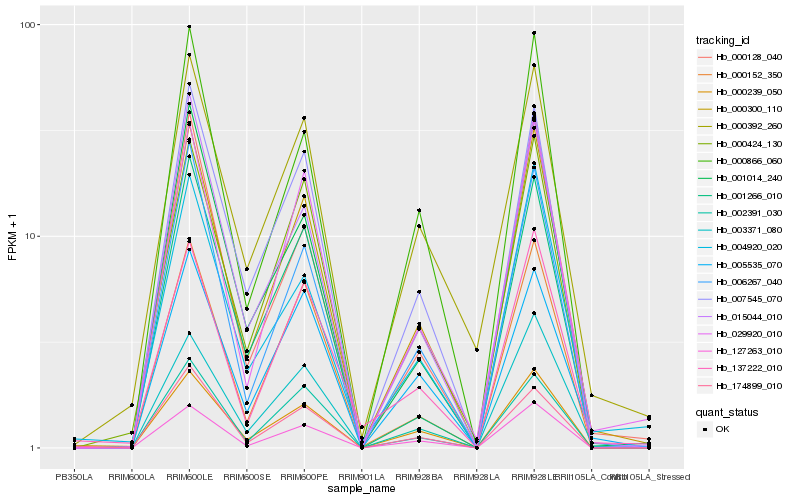
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 2 |
Hb_127263_010 |
0.0617696237 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 3 |
Hb_000239_050 |
0.0677639586 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 4 |
Hb_029920_010 |
0.0685410036 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 5 |
Hb_007545_070 |
0.0733477929 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 6 |
Hb_000866_060 |
0.0781143995 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 7 |
Hb_000152_350 |
0.0788232797 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 8 |
Hb_000424_130 |
0.0788330889 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 9 |
Hb_015044_010 |
0.0788581766 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 10 |
Hb_006267_040 |
0.0795178586 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000128_040 |
0.0861679616 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001014_240 |
0.0882639015 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 13 |
Hb_003371_080 |
0.0914948287 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 14 |
Hb_004920_020 |
0.0961749776 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 15 |
Hb_137222_010 |
0.0969001571 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 16 |
Hb_005535_070 |
0.0990062206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_174899_010 |
0.1006787444 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 18 |
Hb_002391_030 |
0.102566371 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 19 |
Hb_000392_260 |
0.1026135844 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 20 |
Hb_001266_010 |
0.1027346146 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |