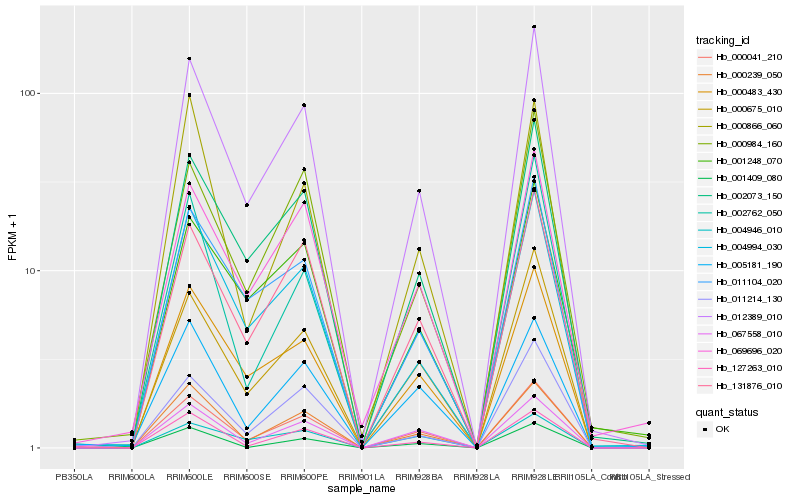
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_210 |
0.0 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 2 |
Hb_012389_010 |
0.0554123259 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 3 |
Hb_067558_010 |
0.0648077804 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 4 |
Hb_000675_010 |
0.0673263093 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 5 |
Hb_131876_010 |
0.0711728196 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000239_050 |
0.0745680603 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 7 |
Hb_000483_430 |
0.0798775756 |
- |
- |
- |
| 8 |
Hb_002073_150 |
0.0802018868 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 9 |
Hb_005181_190 |
0.090561214 |
- |
- |
PREDICTED: uncharacterized protein LOC105643550 [Jatropha curcas] |
| 10 |
Hb_127263_010 |
0.0923980543 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 11 |
Hb_069696_020 |
0.0973646314 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001409_080 |
0.1010063267 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 13 |
Hb_011104_020 |
0.1027538677 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 14 |
Hb_004994_030 |
0.1056505701 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 15 |
Hb_000984_160 |
0.1063910153 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_004946_010 |
0.1103527386 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 17 |
Hb_011214_130 |
0.1131945191 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 18 |
Hb_002762_050 |
0.1166186631 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 19 |
Hb_000866_060 |
0.1166334859 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 20 |
Hb_001248_070 |
0.118722151 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |