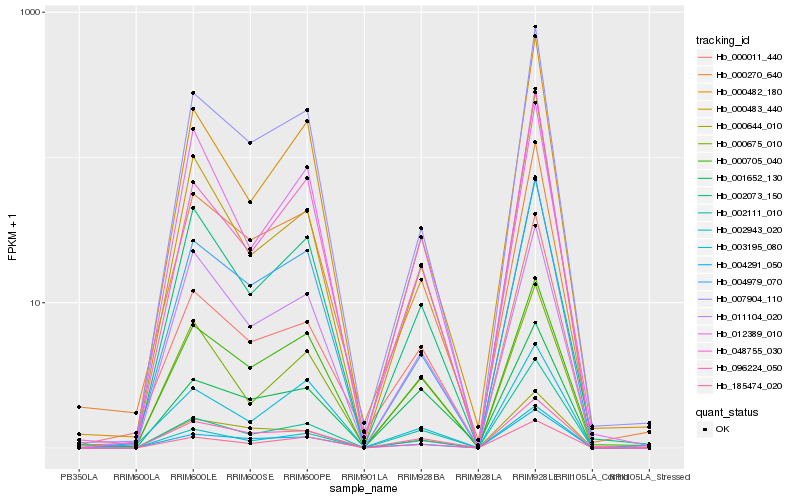
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002943_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 2 |
Hb_000705_040 |
0.0743486252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000675_010 |
0.0834997753 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 4 |
Hb_000270_640 |
0.0982228918 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 5 |
Hb_004979_070 |
0.09900309 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 6 |
Hb_002073_150 |
0.1019808915 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 7 |
Hb_007904_110 |
0.1047857338 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_185474_020 |
0.1048856173 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 9 |
Hb_000011_440 |
0.1052933461 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 10 |
Hb_000644_010 |
0.1061400047 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001652_130 |
0.1071688501 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 12 |
Hb_048755_030 |
0.1084755465 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_011104_020 |
0.1086904984 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 14 |
Hb_000483_440 |
0.1114796034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000482_180 |
0.1116125242 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 16 |
Hb_003195_080 |
0.1129646439 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 17 |
Hb_096224_050 |
0.1138911525 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 18 |
Hb_004291_050 |
0.1147547823 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 19 |
Hb_002111_010 |
0.1151563053 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 20 |
Hb_012389_010 |
0.1161100681 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |