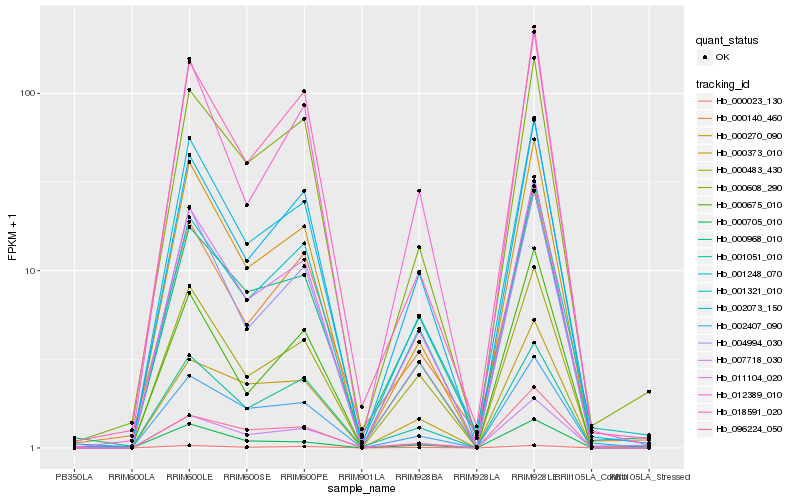
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011104_020 |
0.0 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 2 |
Hb_002073_150 |
0.0489993042 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 3 |
Hb_000483_430 |
0.0527526795 |
- |
- |
- |
| 4 |
Hb_001321_010 |
0.0589406697 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 5 |
Hb_000270_090 |
0.0629571475 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_004994_030 |
0.0660190913 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 7 |
Hb_012389_010 |
0.0683530853 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 8 |
Hb_002407_090 |
0.0743672316 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 9 |
Hb_001248_070 |
0.0752906354 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 10 |
Hb_000608_290 |
0.0840668812 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 11 |
Hb_007718_030 |
0.0884251084 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_000968_010 |
0.0900983592 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001051_010 |
0.0910004823 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 14 |
Hb_000140_460 |
0.0931850892 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 15 |
Hb_000675_010 |
0.0980443762 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 16 |
Hb_018591_020 |
0.098527308 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000023_130 |
0.0991225142 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 18 |
Hb_096224_050 |
0.0995533596 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 19 |
Hb_000705_010 |
0.1002698424 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 20 |
Hb_000373_010 |
0.1006702304 |
- |
- |
ATP binding protein, putative [Ricinus communis] |