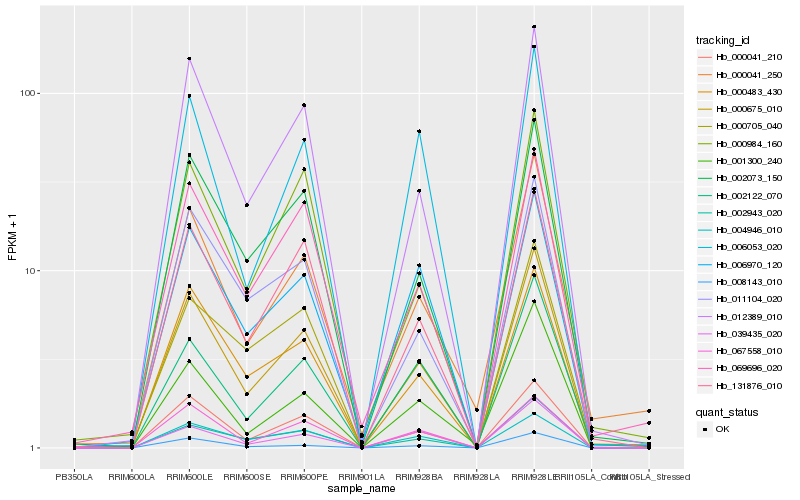
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000675_010 |
0.0 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 2 |
Hb_000041_210 |
0.0673263093 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 3 |
Hb_012389_010 |
0.068716788 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 4 |
Hb_002943_020 |
0.0834997753 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 5 |
Hb_002073_150 |
0.0846105182 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 6 |
Hb_000483_430 |
0.0882283548 |
- |
- |
- |
| 7 |
Hb_131876_010 |
0.092128414 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_011104_020 |
0.0980443762 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 9 |
Hb_067558_010 |
0.0995285111 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_069696_020 |
0.1027749374 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_008143_010 |
0.102789125 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 12 |
Hb_002122_070 |
0.1062050541 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001300_240 |
0.1065899412 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000041_250 |
0.1066258074 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 15 |
Hb_006970_120 |
0.1067113279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000984_160 |
0.1068006237 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000705_040 |
0.109662091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006053_020 |
0.1100047929 |
- |
- |
CAP [Theobroma cacao] |
| 19 |
Hb_004946_010 |
0.1112146098 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 20 |
Hb_039435_020 |
0.1112752253 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |