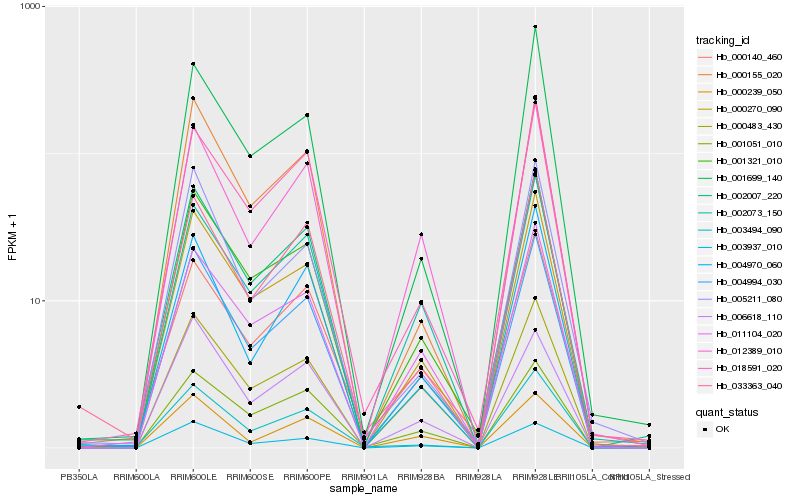
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_030 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 2 |
Hb_000270_090 |
0.0467182893 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 3 |
Hb_001321_010 |
0.0469457218 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 4 |
Hb_003937_010 |
0.0656699639 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 5 |
Hb_011104_020 |
0.0660190913 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 6 |
Hb_012389_010 |
0.0660559647 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 7 |
Hb_002073_150 |
0.0759387879 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 8 |
Hb_002007_220 |
0.0767010203 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000155_020 |
0.0814011554 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000483_430 |
0.0823106973 |
- |
- |
- |
| 11 |
Hb_018591_020 |
0.0844255192 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_004970_060 |
0.0917637568 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 13 |
Hb_000140_460 |
0.0923689305 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 14 |
Hb_005211_080 |
0.0925135546 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 15 |
Hb_001051_010 |
0.0927831206 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 16 |
Hb_001699_140 |
0.0931723333 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006618_110 |
0.0940422485 |
- |
- |
- |
| 18 |
Hb_033363_040 |
0.09458521 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 19 |
Hb_000239_050 |
0.0962659854 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 20 |
Hb_003494_090 |
0.0975437211 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |