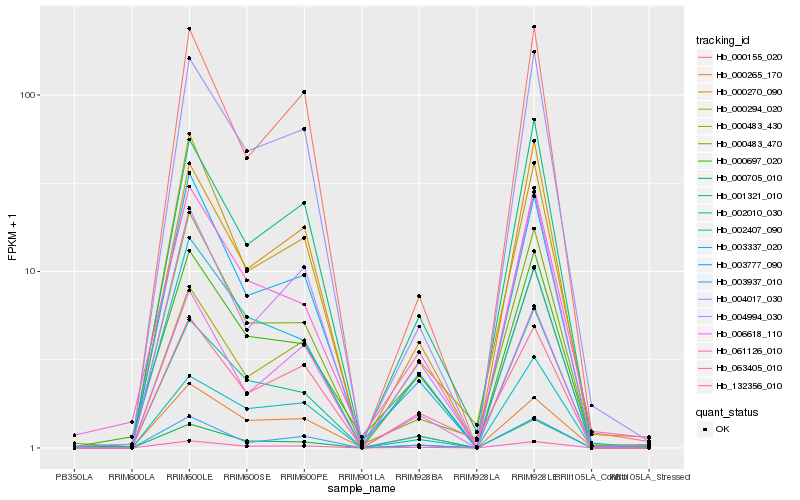
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063405_010 |
0.0 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 2 |
Hb_003937_010 |
0.0678199476 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 3 |
Hb_003777_090 |
0.0678552489 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 4 |
Hb_000705_010 |
0.0738933441 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 5 |
Hb_001321_010 |
0.0899981232 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 6 |
Hb_000270_090 |
0.0912273587 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_000697_020 |
0.0914016838 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_006618_110 |
0.0918435238 |
- |
- |
- |
| 9 |
Hb_003337_020 |
0.0959293584 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 10 |
Hb_000294_020 |
0.0965154644 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 11 |
Hb_004017_030 |
0.0991416114 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 12 |
Hb_004994_030 |
0.1007694298 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 13 |
Hb_132356_010 |
0.1015290449 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_000483_470 |
0.1016131666 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_061126_010 |
0.1028018245 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_000265_170 |
0.1053947709 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000155_020 |
0.1072485725 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002010_030 |
0.1084359043 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_002407_090 |
0.1087546006 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 20 |
Hb_000483_430 |
0.110773241 |
- |
- |
- |