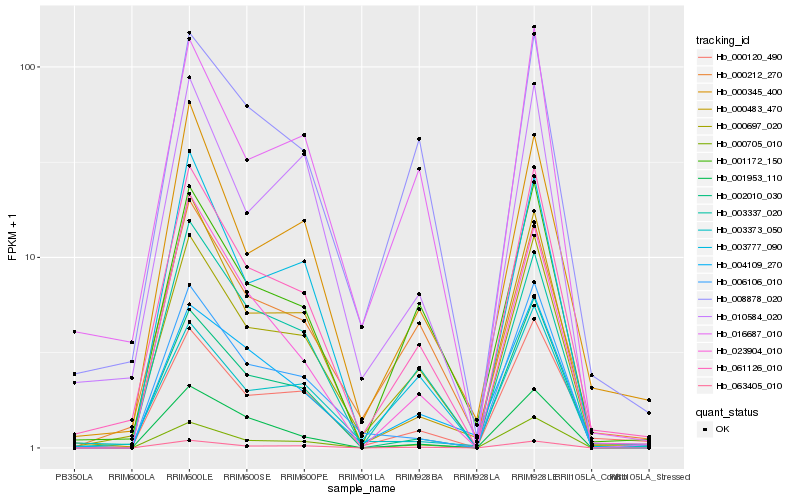
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061126_010 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 2 |
Hb_000697_020 |
0.0569100949 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_000120_490 |
0.0958631612 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 4 |
Hb_000705_010 |
0.0989619897 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 5 |
Hb_063405_010 |
0.1028018245 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 6 |
Hb_002010_030 |
0.1028696331 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_001172_150 |
0.1036068638 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 8 |
Hb_000483_470 |
0.109344387 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003337_020 |
0.111242863 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 10 |
Hb_000212_270 |
0.1151703674 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 11 |
Hb_016687_010 |
0.1152572781 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 12 |
Hb_000345_400 |
0.117678423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003777_090 |
0.1211364336 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 14 |
Hb_003373_050 |
0.1220985903 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 15 |
Hb_001953_110 |
0.1235482891 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 16 |
Hb_004109_270 |
0.124466324 |
transcription factor |
TF Family: NAC |
transcription activator NAC1 family protein [Populus trichocarpa] |
| 17 |
Hb_023904_010 |
0.1255079001 |
- |
- |
- |
| 18 |
Hb_008878_020 |
0.1268822148 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 19 |
Hb_006106_010 |
0.1273779282 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_010584_020 |
0.1278965526 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |