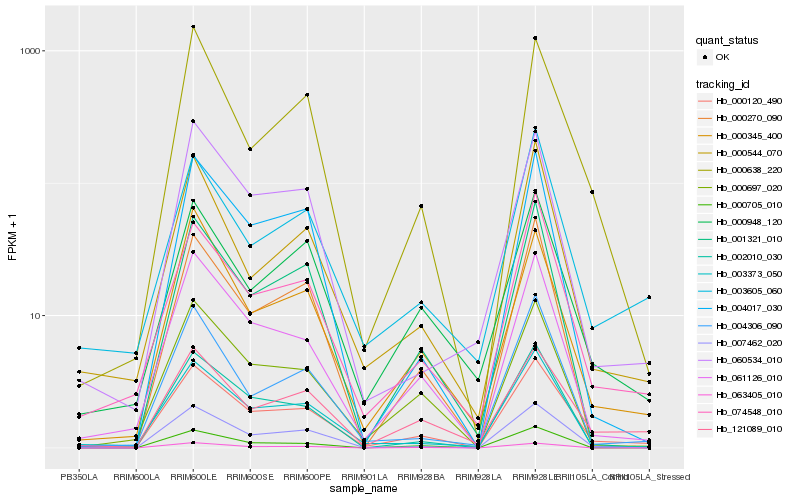
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_490 |
0.0 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 2 |
Hb_061126_010 |
0.0958631612 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_074548_010 |
0.1018733714 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_007462_020 |
0.1041705639 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_121089_010 |
0.1077150798 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 6 |
Hb_002010_030 |
0.11086934 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000697_020 |
0.1128182656 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_003373_050 |
0.1157669523 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 9 |
Hb_004017_030 |
0.1203160971 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 10 |
Hb_000345_400 |
0.1208597897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000638_220 |
0.1223277388 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001321_010 |
0.1224433213 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 13 |
Hb_000544_070 |
0.1227217257 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 14 |
Hb_000948_120 |
0.1234611022 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000270_090 |
0.1253479519 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_004306_090 |
0.1266393989 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 17 |
Hb_063405_010 |
0.1285041399 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 18 |
Hb_000705_010 |
0.1288038966 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 19 |
Hb_060534_010 |
0.1299330986 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 20 |
Hb_003605_060 |
0.1303727995 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |