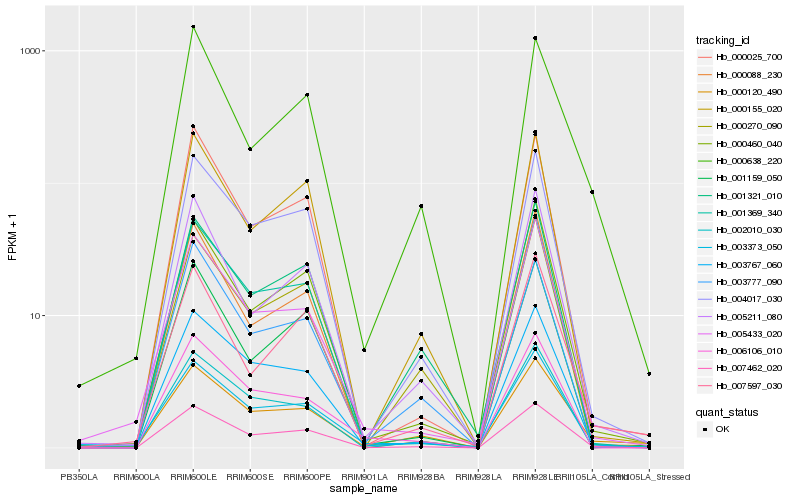
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007462_020 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_004017_030 |
0.0625619549 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 3 |
Hb_003767_060 |
0.0783634189 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 4 |
Hb_001369_340 |
0.0804384387 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 5 |
Hb_000460_040 |
0.0855368182 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 6 |
Hb_000155_020 |
0.0857227404 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_700 |
0.0910500275 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 8 |
Hb_005211_080 |
0.0925882065 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 9 |
Hb_002010_030 |
0.0977588356 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_001159_050 |
0.0982966803 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 11 |
Hb_000270_090 |
0.1023276078 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 12 |
Hb_000120_490 |
0.1041705639 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 13 |
Hb_001321_010 |
0.1041749866 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 14 |
Hb_005433_020 |
0.104826347 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 15 |
Hb_000088_230 |
0.1052282814 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 16 |
Hb_003373_050 |
0.1076932818 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 17 |
Hb_003777_090 |
0.108533296 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 18 |
Hb_007597_030 |
0.1086432257 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000638_220 |
0.1087007273 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006106_010 |
0.1087510161 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |