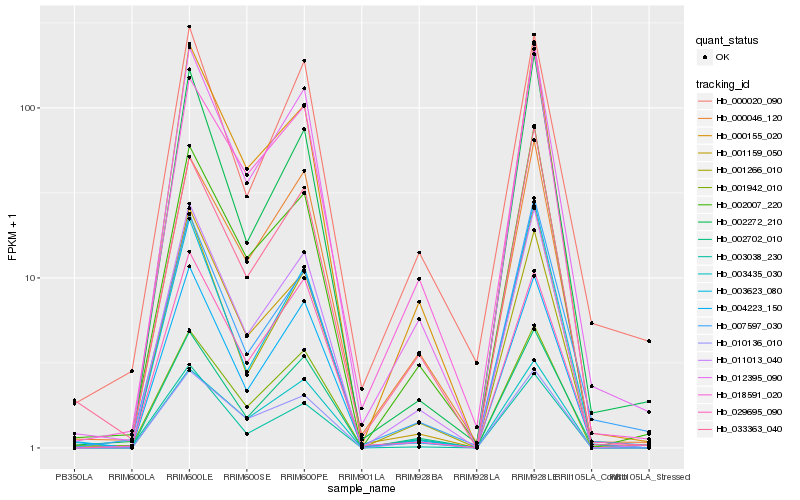
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_090 |
0.0 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 2 |
Hb_011013_040 |
0.0477031268 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 3 |
Hb_002702_010 |
0.050121799 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 4 |
Hb_000155_020 |
0.0594105817 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001942_010 |
0.0680178984 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 6 |
Hb_004223_150 |
0.0686399715 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 7 |
Hb_001159_050 |
0.0703102952 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 8 |
Hb_002007_220 |
0.0749876718 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000046_120 |
0.075254577 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_010136_010 |
0.0797320717 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 11 |
Hb_001266_010 |
0.082232537 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 12 |
Hb_018591_020 |
0.0884508134 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_029695_090 |
0.0886851899 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 14 |
Hb_007597_030 |
0.0889897835 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000020_090 |
0.0890033165 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 16 |
Hb_002272_210 |
0.0890731076 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003623_080 |
0.0894368752 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 18 |
Hb_003435_030 |
0.0900104935 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_033363_040 |
0.0909287577 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 20 |
Hb_003038_230 |
0.0935350048 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |