| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
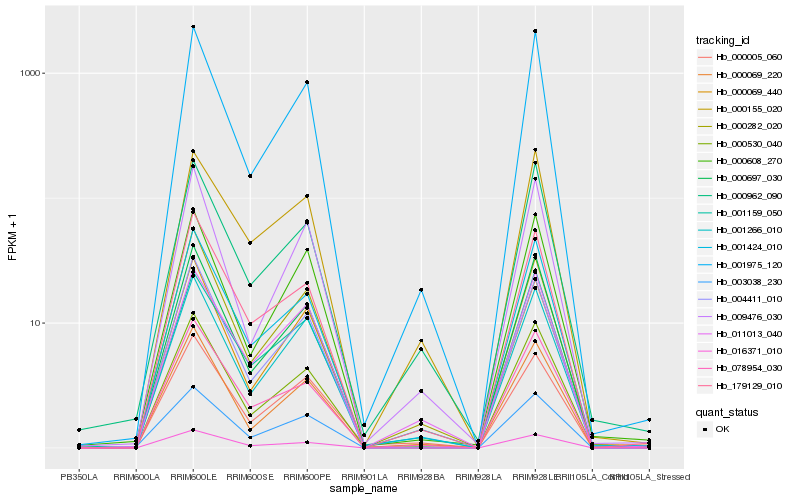
Hb_003038_230 |
0.0 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 2 |
Hb_001266_010 |
0.0682547057 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 3 |
Hb_001975_120 |
0.0700124828 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 4 |
Hb_011013_040 |
0.0712277941 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 5 |
Hb_001424_010 |
0.0735397865 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000697_030 |
0.0754020719 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 7 |
Hb_001159_050 |
0.0779358876 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 8 |
Hb_000608_270 |
0.0784473646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004411_010 |
0.0795489453 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 10 |
Hb_000530_040 |
0.0796242097 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000005_060 |
0.0828857371 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 12 |
Hb_000069_440 |
0.084099305 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 13 |
Hb_179129_010 |
0.0841860341 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 14 |
Hb_000962_090 |
0.0855834065 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 15 |
Hb_016371_010 |
0.0856044543 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_000155_020 |
0.0888380256 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000069_220 |
0.0900702222 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 18 |
Hb_000282_020 |
0.0903744729 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009476_030 |
0.0919294829 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 20 |
Hb_078954_030 |
0.0926087447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |