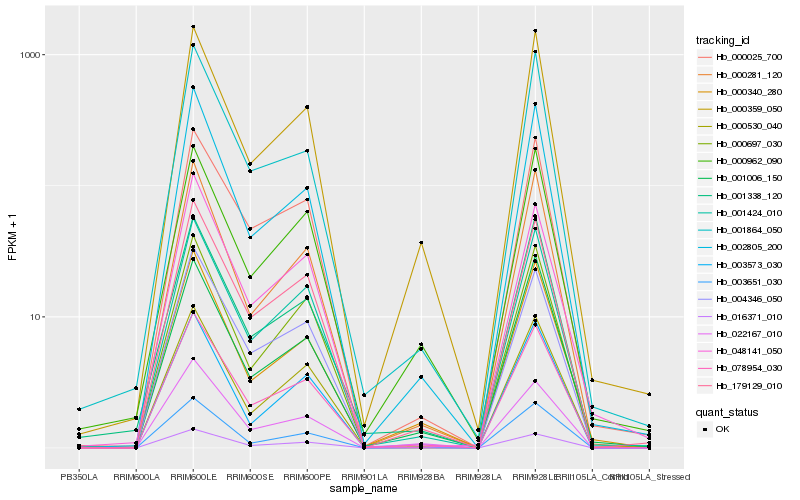
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078954_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001424_010 |
0.0338241543 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_179129_010 |
0.0402706338 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 4 |
Hb_004346_050 |
0.0448120045 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 5 |
Hb_000025_700 |
0.0508915827 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 6 |
Hb_016371_010 |
0.052082953 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 7 |
Hb_000530_040 |
0.057815381 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002805_200 |
0.0579087116 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 9 |
Hb_000281_120 |
0.0621991975 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000359_050 |
0.0627386285 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000340_280 |
0.0630333143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003651_030 |
0.0637638936 |
- |
- |
lyase, putative [Ricinus communis] |
| 13 |
Hb_001864_050 |
0.0703868783 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 14 |
Hb_000697_030 |
0.0715201176 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 15 |
Hb_003573_030 |
0.0748599696 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 16 |
Hb_022167_010 |
0.0758800786 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 17 |
Hb_001006_150 |
0.076606117 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_001338_120 |
0.0782464239 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 19 |
Hb_048141_050 |
0.0784060403 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 20 |
Hb_000962_090 |
0.0796372233 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |