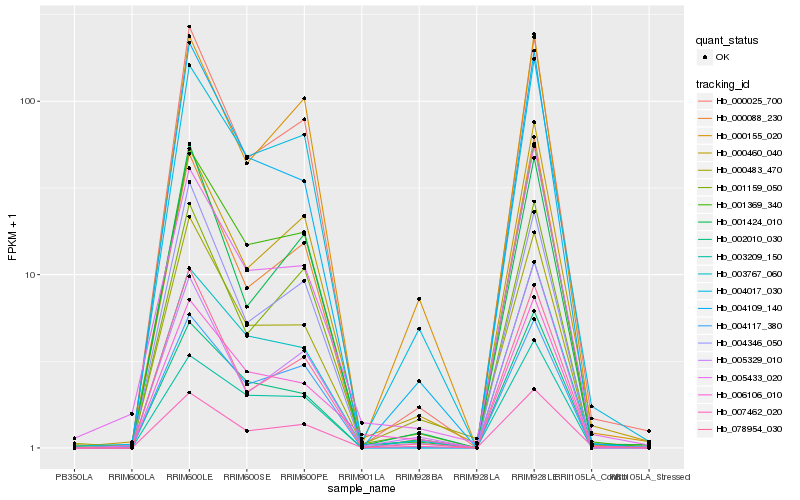
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_340 |
0.0 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 2 |
Hb_003767_060 |
0.0570309188 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 3 |
Hb_000025_700 |
0.0607511572 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 4 |
Hb_004117_380 |
0.0620250895 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 5 |
Hb_004017_030 |
0.065795358 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 6 |
Hb_003209_150 |
0.0665914985 |
- |
- |
CLE9, putative [Ricinus communis] |
| 7 |
Hb_007462_020 |
0.0804384387 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_000088_230 |
0.0817918782 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 9 |
Hb_002010_030 |
0.0882697309 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_004109_140 |
0.0889609722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001159_050 |
0.0906745175 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 12 |
Hb_000460_040 |
0.091432535 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 13 |
Hb_000155_020 |
0.0929139001 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_005433_020 |
0.0981125182 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 15 |
Hb_078954_030 |
0.1007794103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006106_010 |
0.1011819704 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001424_010 |
0.105666586 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000483_470 |
0.107158617 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004346_050 |
0.1077379443 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 20 |
Hb_005329_010 |
0.1114400741 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |