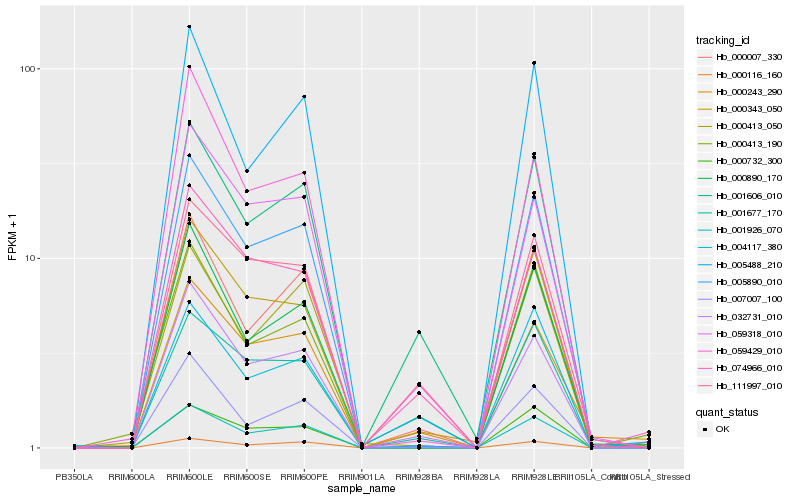
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005890_010 |
0.0 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 2 |
Hb_000116_160 |
0.0733535137 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 3 |
Hb_005488_210 |
0.0848350075 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 4 |
Hb_001926_070 |
0.089298236 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 5 |
Hb_004117_380 |
0.0895554292 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 6 |
Hb_000343_050 |
0.0904272024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_032731_010 |
0.0934936231 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 8 |
Hb_000007_330 |
0.0972571964 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 9 |
Hb_001677_170 |
0.0981088428 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 10 |
Hb_000413_050 |
0.1026882537 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007007_100 |
0.1045387298 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 12 |
Hb_059318_010 |
0.1097831757 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 13 |
Hb_000890_170 |
0.1105002469 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 14 |
Hb_000732_300 |
0.1106265423 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001606_010 |
0.113563466 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000413_190 |
0.1138705741 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 17 |
Hb_059429_010 |
0.1165332647 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 18 |
Hb_000243_290 |
0.11787041 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 19 |
Hb_111997_010 |
0.1210730931 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 20 |
Hb_074966_010 |
0.1248161234 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |