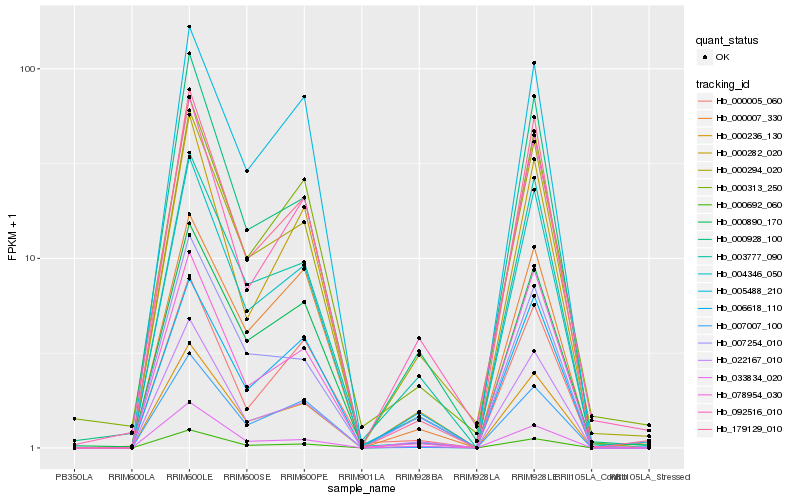
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_130 |
0.0 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 2 |
Hb_022167_010 |
0.0618959553 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 3 |
Hb_000692_060 |
0.0637901615 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000890_170 |
0.0649954325 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 5 |
Hb_003777_090 |
0.0654140203 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 6 |
Hb_000294_020 |
0.0693610594 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 7 |
Hb_179129_010 |
0.0752038499 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 8 |
Hb_004346_050 |
0.0771671027 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 9 |
Hb_000928_100 |
0.0821099991 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_000282_020 |
0.0841980961 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007254_010 |
0.0857744995 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 12 |
Hb_005488_210 |
0.0873285585 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_092516_010 |
0.090933901 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 14 |
Hb_000007_330 |
0.0911664645 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 15 |
Hb_006618_110 |
0.0919235831 |
- |
- |
- |
| 16 |
Hb_000005_060 |
0.0941061904 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 17 |
Hb_033834_020 |
0.094215167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000313_250 |
0.0942512334 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 19 |
Hb_007007_100 |
0.0943747469 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 20 |
Hb_078954_030 |
0.0956102743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |