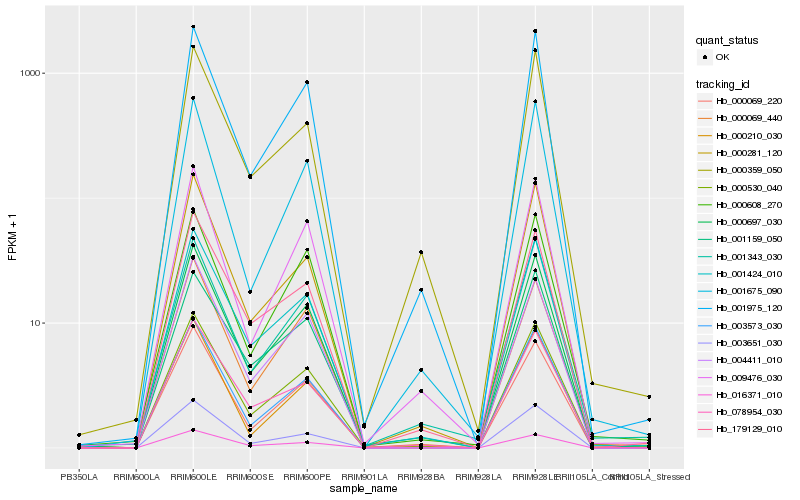
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000530_040 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001424_010 |
0.0311014148 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000697_030 |
0.0401131586 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 4 |
Hb_003573_030 |
0.0461365446 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 5 |
Hb_001975_120 |
0.0473233934 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 6 |
Hb_000069_220 |
0.0545603815 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 7 |
Hb_078954_030 |
0.057815381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000281_120 |
0.0594305665 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_016371_010 |
0.0617855024 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_179129_010 |
0.0626423678 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 11 |
Hb_003651_030 |
0.0628425806 |
- |
- |
lyase, putative [Ricinus communis] |
| 12 |
Hb_004411_010 |
0.0653293454 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 13 |
Hb_001343_030 |
0.0672261541 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000069_440 |
0.0693503186 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 15 |
Hb_009476_030 |
0.0743718416 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 16 |
Hb_000359_050 |
0.0748049627 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001675_090 |
0.0748866749 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000608_270 |
0.0753731847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001159_050 |
0.0757347701 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 20 |
Hb_000210_030 |
0.0759304303 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |