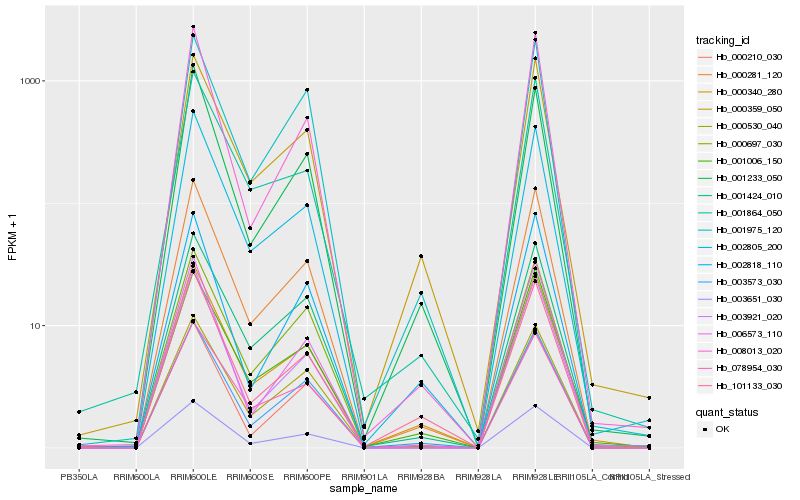
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003651_030 |
0.0 |
- |
- |
lyase, putative [Ricinus communis] |
| 2 |
Hb_000281_120 |
0.0168707608 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_003573_030 |
0.0393236261 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 4 |
Hb_002805_200 |
0.043043009 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 5 |
Hb_000210_030 |
0.0543339268 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 6 |
Hb_000340_280 |
0.0554271718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000359_050 |
0.0587776253 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001424_010 |
0.0612560563 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000530_040 |
0.0628425806 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_078954_030 |
0.0637638936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003921_020 |
0.0645072465 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 12 |
Hb_001006_150 |
0.0647158056 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_101133_030 |
0.0661126923 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_008013_020 |
0.0666214479 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000697_030 |
0.0676150156 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 16 |
Hb_001233_050 |
0.0679382923 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_006573_110 |
0.0681739163 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 18 |
Hb_002818_110 |
0.0690335303 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 19 |
Hb_001975_120 |
0.0697272372 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 20 |
Hb_001864_050 |
0.0719796401 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |