| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
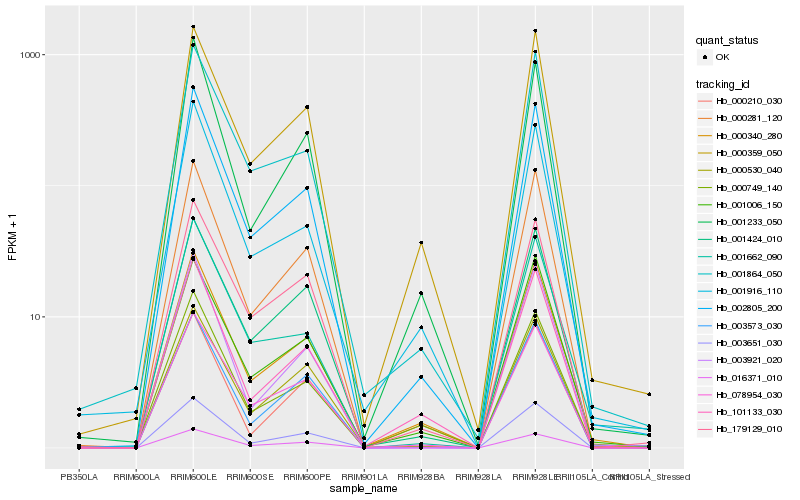
Hb_002805_200 |
0.0 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 2 |
Hb_000281_120 |
0.0401895872 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_003651_030 |
0.043043009 |
- |
- |
lyase, putative [Ricinus communis] |
| 4 |
Hb_000340_280 |
0.0508088511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001864_050 |
0.0564206373 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 6 |
Hb_078954_030 |
0.0579087116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001233_050 |
0.0627407948 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_003921_020 |
0.0652369831 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 9 |
Hb_003573_030 |
0.0691810567 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 10 |
Hb_000359_050 |
0.0701653875 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001424_010 |
0.0705138081 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001916_110 |
0.0723551947 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 13 |
Hb_101133_030 |
0.0728440855 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_179129_010 |
0.0739855401 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 15 |
Hb_001662_090 |
0.0758311555 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000530_040 |
0.0762502336 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000749_140 |
0.0767205231 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 18 |
Hb_001006_150 |
0.0778517038 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000210_030 |
0.0819391967 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 20 |
Hb_016371_010 |
0.0827027603 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |