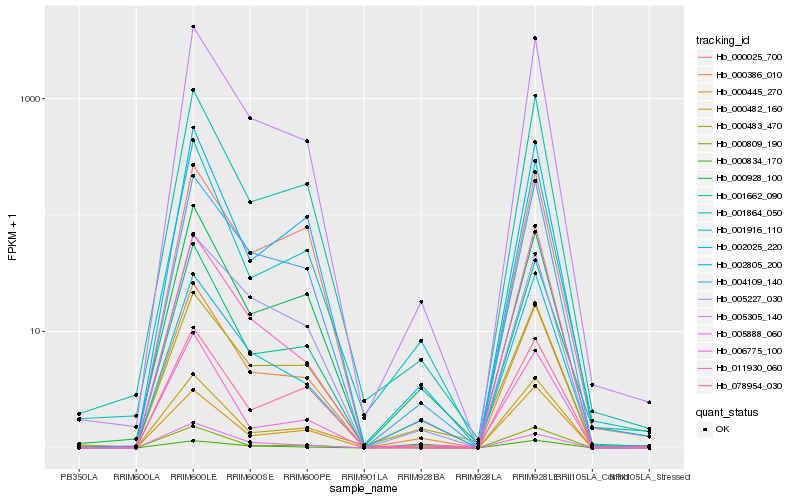
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_140 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_004109_140 |
0.050858608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000386_010 |
0.0565563554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001864_050 |
0.0691321424 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 5 |
Hb_002025_220 |
0.0764237833 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001662_090 |
0.0810903004 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000834_170 |
0.0816002284 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 8 |
Hb_005227_030 |
0.0929236976 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000482_160 |
0.0942853548 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 10 |
Hb_002805_200 |
0.0990941296 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 11 |
Hb_000483_470 |
0.099223954 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_011930_060 |
0.1006477579 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_005888_060 |
0.1030027111 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 14 |
Hb_000445_270 |
0.1050489341 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000928_100 |
0.1054306618 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_000809_190 |
0.1062209688 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 17 |
Hb_078954_030 |
0.1063083868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001916_110 |
0.1065969793 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 19 |
Hb_006775_100 |
0.1090392675 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000025_700 |
0.1148608541 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |