Hb_001864_050
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig1864: 43095-44455 |
| Sequence |   |
Annotation
kegg
| ID | pop:POPTR_0011s03390g |
|---|---|
| description | POPTRDRAFT_834003; Oxygen-evolving enhancer protein 3-1 |
nr
| ID | AAV74404.1 |
|---|---|
| description | chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
swissprot
| ID | Q41932 |
|---|---|
| description | Oxygen-evolving enhancer protein 3-2, chloroplastic OS=Arabidopsis thaliana GN=PSBQ2 PE=1 SV=2 |
trembl
| ID | Q5PYQ5 |
|---|---|
| description | Chloroplast oxygen-evolving enhancer protein OS=Manihot esculenta PE=2 SV=1 |
Gene Ontology
| ID | GO:0009654 |
|---|---|
| description | oxygen-evolving enhancer protein 3- chloroplastic-like |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_18404: 43207-44378 |
|---|---|
| cDNA (Sanger) (ID:Location) |
001_O22.ab1: 43261-44245 , 002_P14.ab1: 43241-44237 , 007_K21.ab1: 43241-44301 , 009_L12.ab1: 43261-44235 , 011_K01.ab1: 43241-44276 , 012_A22.ab1: 43241-44238 , 015_K09.ab1: 43265-43842 , 017_P13.ab1: 43261-44215 , 020_P03.ab1: 43261-44247 , 022_H03.ab1: 43269-43935 , 023_J07.ab1: 43207-44021 , 024_E02.ab1: 43267-43960 , 025_G14.ab1: 43261-44251 , 026_B21.ab1: 43261-44166 , 026_J13.ab1: 43261-44185 , 029_N19.ab1: 43241-43939 , 033_K10.ab1: 43266-44126 , 037_E19.ab1: 43261-44111 , 042_N21.ab1: 43261-44253 , 043_A18.ab1: 43261-44215 , 044_I10.ab1: 43261-44215 , 045_O19.ab1: 43261-44120 , 046_E23.ab1: 43261-44222 , 053_L17.ab1: 43204-44184 |
Similar expressed genes (Top20)
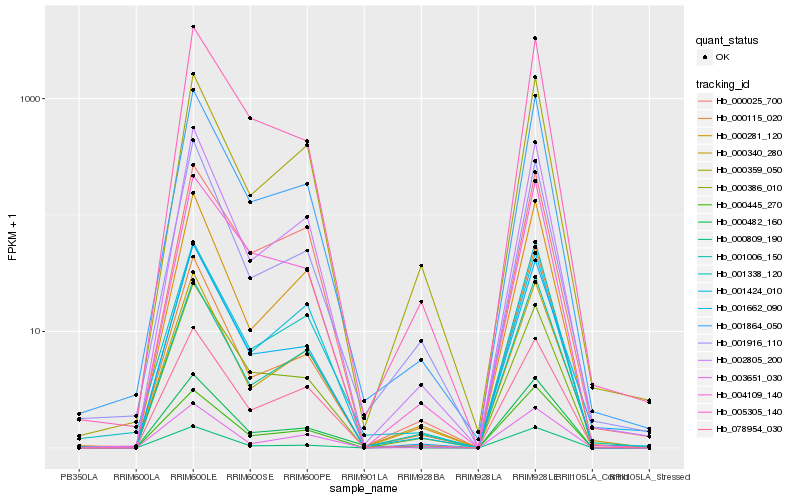
Gene co-expression network
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 1.85865 | 128.279 | 1190.68 | 184.432 | 0.969542 | 1.52901 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 1.05859 | 0.466303 | 0.185535 | 4.69977 | 1057.25 |

