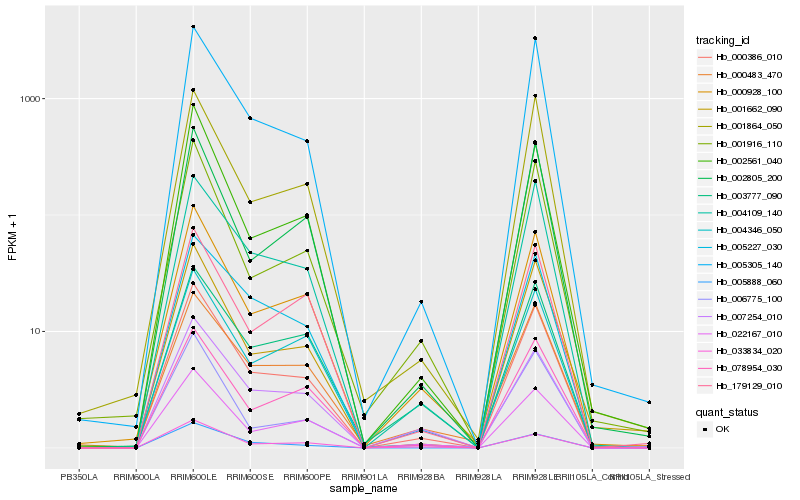
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000386_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005305_140 |
0.0565563554 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_000928_100 |
0.0658896766 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 4 |
Hb_004109_140 |
0.0769452655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_022167_010 |
0.0781005909 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 6 |
Hb_005888_060 |
0.0848902976 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 7 |
Hb_001662_090 |
0.0871816529 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001864_050 |
0.0874745786 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 9 |
Hb_005227_030 |
0.0876128685 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002561_040 |
0.0877135841 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 11 |
Hb_002805_200 |
0.0885038744 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 12 |
Hb_001916_110 |
0.0915689091 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 13 |
Hb_007254_010 |
0.0926106047 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 14 |
Hb_004346_050 |
0.0929118367 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 15 |
Hb_078954_030 |
0.0939888115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000483_470 |
0.0963287274 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_179129_010 |
0.098088324 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 18 |
Hb_033834_020 |
0.0987454849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006775_100 |
0.1030500576 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_003777_090 |
0.1034103913 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |