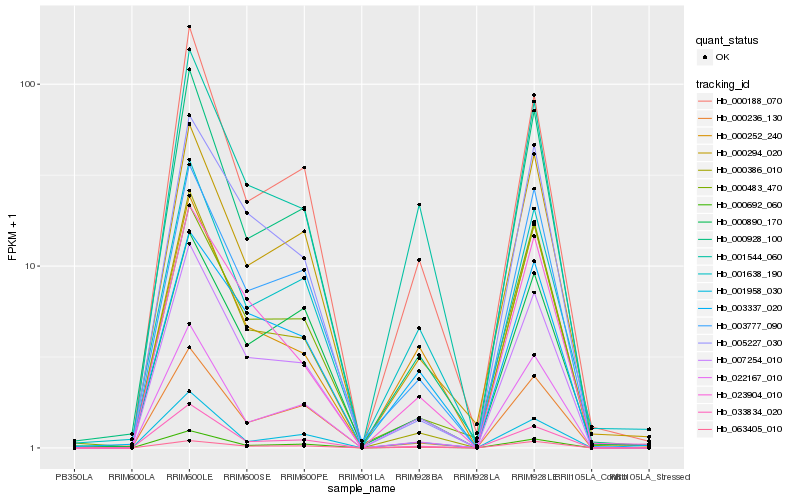
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007254_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 2 |
Hb_000692_060 |
0.0499370007 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 3 |
Hb_033834_020 |
0.054906783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000188_070 |
0.0722554622 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000928_100 |
0.0818873455 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_022167_010 |
0.0842452948 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 7 |
Hb_003777_090 |
0.0846835945 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 8 |
Hb_000236_130 |
0.0857744995 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 9 |
Hb_000386_010 |
0.0926106047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000294_020 |
0.0946129434 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 11 |
Hb_001958_030 |
0.1007792455 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 12 |
Hb_023904_010 |
0.1044299745 |
- |
- |
- |
| 13 |
Hb_005227_030 |
0.1056664781 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001544_060 |
0.1064792489 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001638_190 |
0.1109766524 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000483_470 |
0.1118725054 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000890_170 |
0.1145514016 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 18 |
Hb_003337_020 |
0.1145955955 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 19 |
Hb_000252_240 |
0.1190741229 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_063405_010 |
0.1199065273 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |