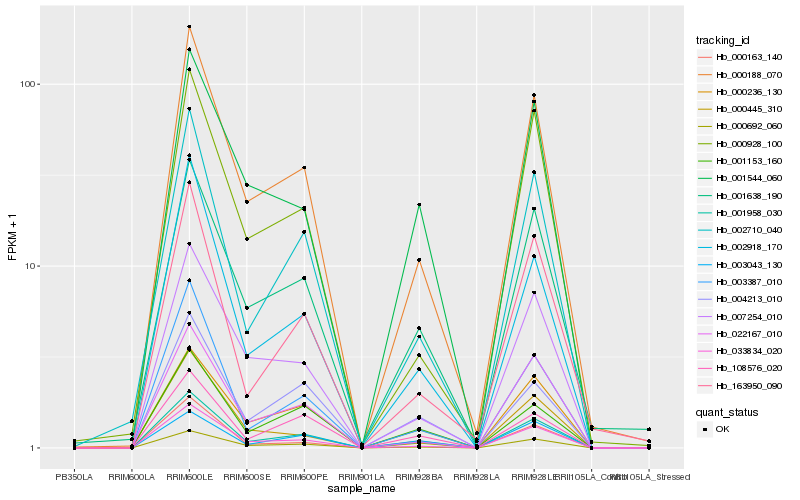
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001958_030 |
0.0 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 2 |
Hb_000188_070 |
0.0521309654 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_000692_060 |
0.062189567 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_002710_040 |
0.0725958513 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 5 |
Hb_033834_020 |
0.075664357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001638_190 |
0.0935155788 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003387_010 |
0.0950569147 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 8 |
Hb_002918_170 |
0.0985433229 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 9 |
Hb_022167_010 |
0.1007358509 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 10 |
Hb_007254_010 |
0.1007792455 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 11 |
Hb_000163_140 |
0.1033381769 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 12 |
Hb_108576_020 |
0.1038231501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001153_160 |
0.1064755233 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 14 |
Hb_004213_010 |
0.1083588032 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001544_060 |
0.1121462098 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_163950_090 |
0.1127380447 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_003043_130 |
0.1130821913 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 18 |
Hb_000928_100 |
0.1131130198 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_000236_130 |
0.1141372924 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 20 |
Hb_000445_310 |
0.1147369157 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |