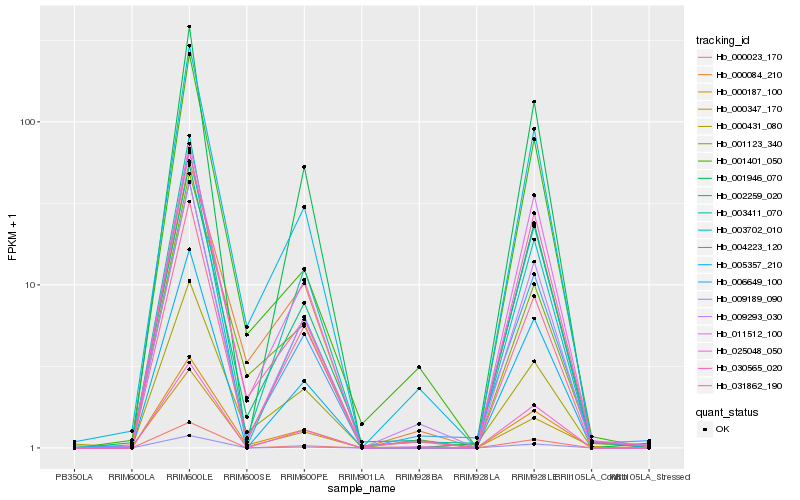
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_120 |
0.0 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000187_100 |
0.0555317188 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 3 |
Hb_009293_030 |
0.0677294584 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 4 |
Hb_030565_020 |
0.0699570032 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 5 |
Hb_002259_020 |
0.0725308285 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 6 |
Hb_006649_100 |
0.0750374178 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_001401_050 |
0.0801026144 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031862_190 |
0.0840815207 |
- |
- |
- |
| 9 |
Hb_003411_070 |
0.0845060809 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 10 |
Hb_000084_210 |
0.0857641401 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 11 |
Hb_003702_010 |
0.0883905691 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 12 |
Hb_005357_210 |
0.0897456954 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 13 |
Hb_025048_050 |
0.0929048718 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 14 |
Hb_000023_170 |
0.0933756259 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 15 |
Hb_000431_080 |
0.0938669145 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 16 |
Hb_011512_100 |
0.0942142922 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001946_070 |
0.0946709866 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000347_170 |
0.0947241816 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_009189_090 |
0.0960450805 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 20 |
Hb_001123_340 |
0.0966033261 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |