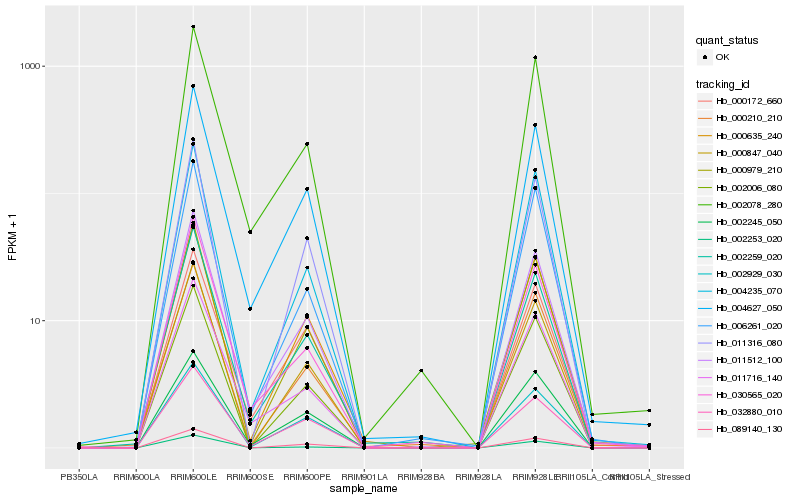
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011512_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004627_050 |
0.0246597917 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 3 |
Hb_002259_020 |
0.0500100453 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 4 |
Hb_002078_280 |
0.0513029028 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 5 |
Hb_011716_140 |
0.0590898482 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 6 |
Hb_000979_210 |
0.0593334153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_030565_020 |
0.0612761495 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 8 |
Hb_000172_660 |
0.0672507599 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_002245_050 |
0.0682086039 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000847_040 |
0.0689966833 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 11 |
Hb_002929_030 |
0.0695180263 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 12 |
Hb_011316_080 |
0.0713413801 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_000635_240 |
0.0715446278 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 14 |
Hb_000210_210 |
0.0727359711 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002006_080 |
0.07503063 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 16 |
Hb_004235_070 |
0.0751293578 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 17 |
Hb_089140_130 |
0.076798975 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 18 |
Hb_002253_020 |
0.0776401993 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 19 |
Hb_006261_020 |
0.078097825 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 20 |
Hb_032880_010 |
0.0784603884 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |