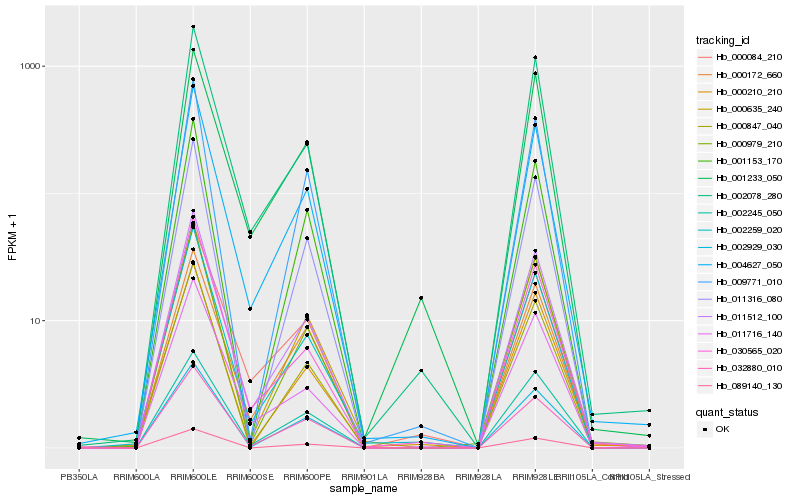
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004627_050 |
0.0 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 2 |
Hb_011512_100 |
0.0246597917 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002259_020 |
0.0500913719 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 4 |
Hb_002078_280 |
0.0513883427 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 5 |
Hb_000172_660 |
0.0536453004 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_002245_050 |
0.0608359317 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002929_030 |
0.0641614139 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 8 |
Hb_011716_140 |
0.0653459824 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 9 |
Hb_000635_240 |
0.0672213277 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 10 |
Hb_000979_210 |
0.0689825747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_032880_010 |
0.0707237024 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 12 |
Hb_011316_080 |
0.0745585941 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_000084_210 |
0.0758941355 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 14 |
Hb_030565_020 |
0.0761636048 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 15 |
Hb_000210_210 |
0.0762248056 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009771_010 |
0.0774834046 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 17 |
Hb_000847_040 |
0.0776502432 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 18 |
Hb_001153_170 |
0.0780320237 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 19 |
Hb_089140_130 |
0.0784830519 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 20 |
Hb_001233_050 |
0.0793114545 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |