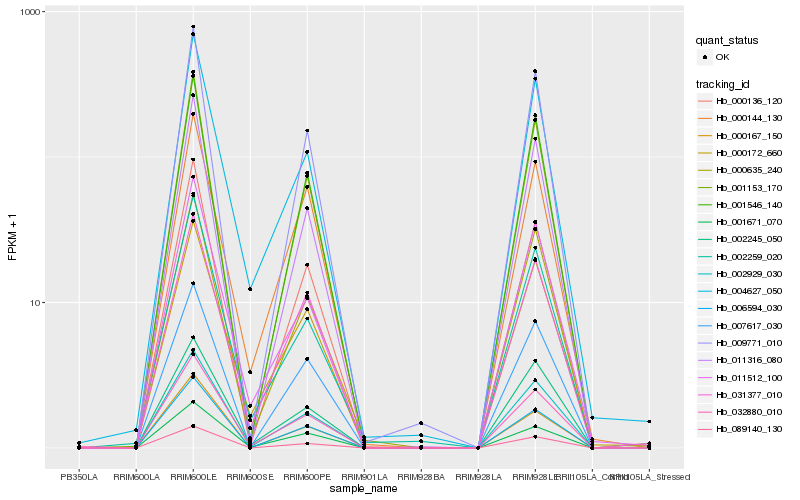
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032880_010 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 2 |
Hb_031377_010 |
0.0633371782 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 3 |
Hb_001671_070 |
0.0634163641 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000172_660 |
0.0641660311 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 5 |
Hb_002929_030 |
0.0647082149 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 6 |
Hb_001153_170 |
0.0685711543 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 7 |
Hb_004627_050 |
0.0707237024 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 8 |
Hb_006594_030 |
0.0720158344 |
- |
- |
- |
| 9 |
Hb_007617_030 |
0.07226318 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 10 |
Hb_009771_010 |
0.0726857088 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 11 |
Hb_089140_130 |
0.0746310099 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 12 |
Hb_000136_120 |
0.0756548096 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 13 |
Hb_001546_140 |
0.0760669044 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 14 |
Hb_000144_130 |
0.0775362247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011512_100 |
0.0784603884 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000635_240 |
0.0791435299 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 17 |
Hb_011316_080 |
0.0798565331 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_000167_150 |
0.0811180715 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 19 |
Hb_002245_050 |
0.0822262072 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_002259_020 |
0.0850726019 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |