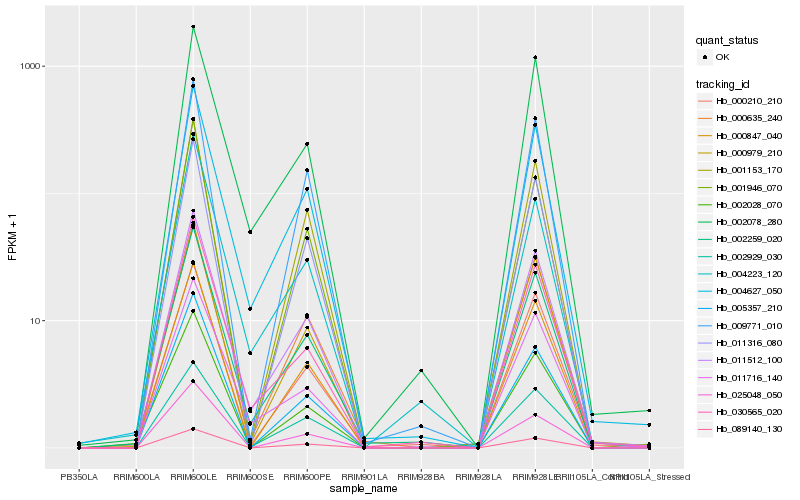
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 2 |
Hb_011512_100 |
0.0500100453 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004627_050 |
0.0500913719 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 4 |
Hb_030565_020 |
0.0630163703 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 5 |
Hb_004223_120 |
0.0725308285 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002078_280 |
0.0735745049 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 7 |
Hb_000847_040 |
0.0736618774 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 8 |
Hb_002929_030 |
0.0740885759 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 9 |
Hb_011716_140 |
0.0767540613 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 10 |
Hb_000210_210 |
0.0780242704 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002028_070 |
0.0784564824 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 12 |
Hb_025048_050 |
0.0784611887 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 13 |
Hb_000635_240 |
0.0787745447 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 14 |
Hb_011316_080 |
0.0788964837 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 15 |
Hb_000979_210 |
0.0797722826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001946_070 |
0.0813565553 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_089140_130 |
0.0816082804 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 18 |
Hb_005357_210 |
0.0819964649 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 19 |
Hb_009771_010 |
0.0827036172 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 20 |
Hb_001153_170 |
0.0830029159 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |