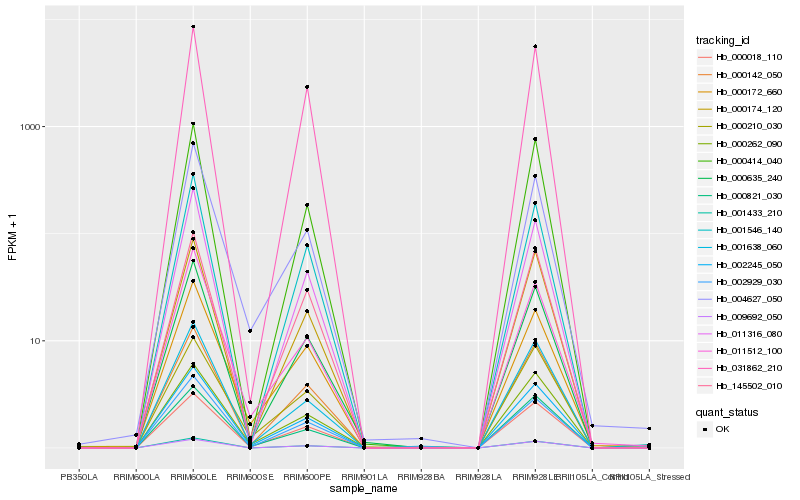
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002245_050 |
0.0 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000635_240 |
0.045225436 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 3 |
Hb_000018_110 |
0.0484323562 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 4 |
Hb_000821_030 |
0.0511470942 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 5 |
Hb_001433_210 |
0.0533750151 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 6 |
Hb_000142_050 |
0.0542072221 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 7 |
Hb_002929_030 |
0.0547302508 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 8 |
Hb_000174_120 |
0.055846667 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 9 |
Hb_000262_090 |
0.0563663394 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 10 |
Hb_000172_660 |
0.0585839355 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000414_040 |
0.0599759054 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 12 |
Hb_004627_050 |
0.0608359317 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 13 |
Hb_145502_010 |
0.0621826841 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 14 |
Hb_001546_140 |
0.063026108 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 15 |
Hb_009692_050 |
0.0640625552 |
- |
- |
- |
| 16 |
Hb_001638_060 |
0.0667704117 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 17 |
Hb_011316_080 |
0.0675882848 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_011512_100 |
0.0682086039 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 19 |
Hb_031862_210 |
0.0691988723 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 20 |
Hb_000210_030 |
0.0692652645 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |