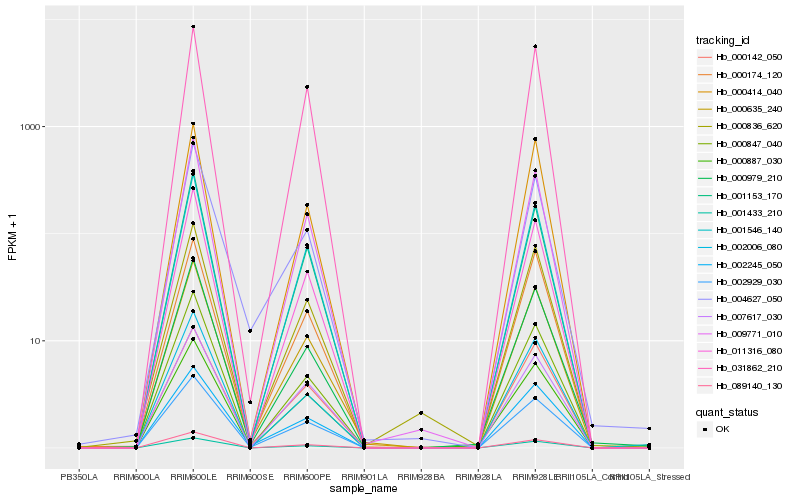
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_240 |
0.0 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 2 |
Hb_011316_080 |
0.0425866552 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_001546_140 |
0.0435382889 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 4 |
Hb_009771_010 |
0.0442068976 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 5 |
Hb_002245_050 |
0.045225436 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001433_210 |
0.0461518015 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 7 |
Hb_002929_030 |
0.0467909208 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 8 |
Hb_089140_130 |
0.0498603883 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 9 |
Hb_001153_170 |
0.0506246803 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 10 |
Hb_000847_040 |
0.0578988148 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 11 |
Hb_000979_210 |
0.0579378034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007617_030 |
0.0581583731 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 13 |
Hb_000414_040 |
0.0606259194 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 14 |
Hb_002006_080 |
0.0608942246 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 15 |
Hb_000836_620 |
0.0621856638 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000887_030 |
0.0637757583 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 17 |
Hb_000174_120 |
0.0647232158 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 18 |
Hb_000142_050 |
0.0664018087 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 19 |
Hb_031862_210 |
0.066406704 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 20 |
Hb_004627_050 |
0.0672213277 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |