| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
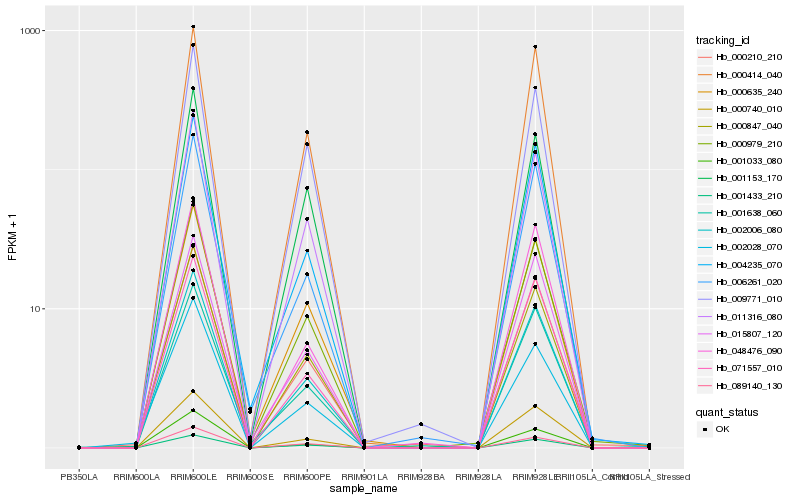
Hb_002006_080 |
0.0 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 2 |
Hb_000847_040 |
0.0277782236 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 3 |
Hb_011316_080 |
0.0421569714 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 4 |
Hb_000979_210 |
0.0428618108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000740_010 |
0.0430006955 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_004235_070 |
0.0504716048 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 7 |
Hb_071557_010 |
0.0547613279 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 8 |
Hb_000210_210 |
0.0557814018 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 9 |
Hb_089140_130 |
0.0568477537 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 10 |
Hb_006261_020 |
0.0597674238 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 11 |
Hb_000635_240 |
0.0608942246 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 12 |
Hb_001033_080 |
0.0633380146 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 13 |
Hb_000414_040 |
0.06341219 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 14 |
Hb_001433_210 |
0.0639041996 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 15 |
Hb_048476_090 |
0.0646504323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009771_010 |
0.06520532 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 17 |
Hb_001638_060 |
0.0660999293 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 18 |
Hb_015807_120 |
0.0678736916 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001153_170 |
0.0689592715 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 20 |
Hb_002028_070 |
0.0700455555 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |