| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
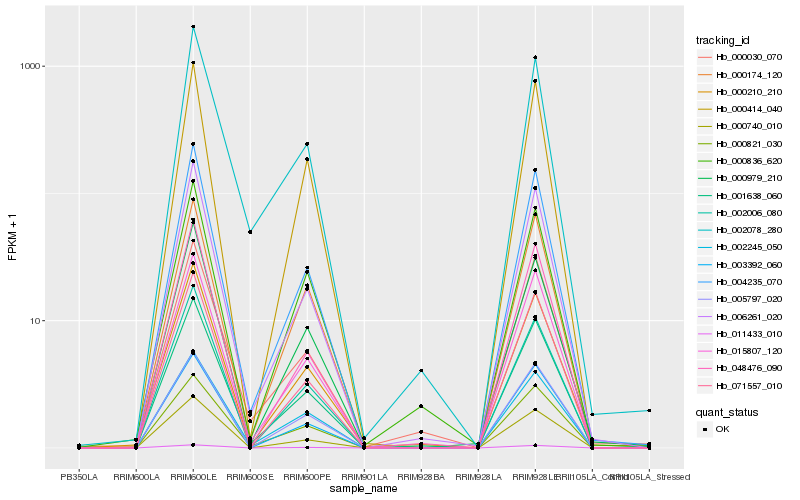
Hb_001638_060 |
0.0 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 2 |
Hb_004235_070 |
0.0445913607 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 3 |
Hb_015807_120 |
0.0460209927 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006261_020 |
0.0469317364 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 5 |
Hb_000210_210 |
0.0532895041 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000821_030 |
0.0542389448 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 7 |
Hb_000030_070 |
0.0565900065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000740_010 |
0.0586263269 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_071557_010 |
0.0593285487 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 10 |
Hb_000414_040 |
0.062977381 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 11 |
Hb_002078_280 |
0.0645123973 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 12 |
Hb_003392_060 |
0.0653124121 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_002006_080 |
0.0660999293 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 14 |
Hb_002245_050 |
0.0667704117 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_048476_090 |
0.0687064538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000836_620 |
0.0699610306 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000979_210 |
0.0701701016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005797_020 |
0.0704327549 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 19 |
Hb_011433_010 |
0.0704340809 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 20 |
Hb_000174_120 |
0.0706933723 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |