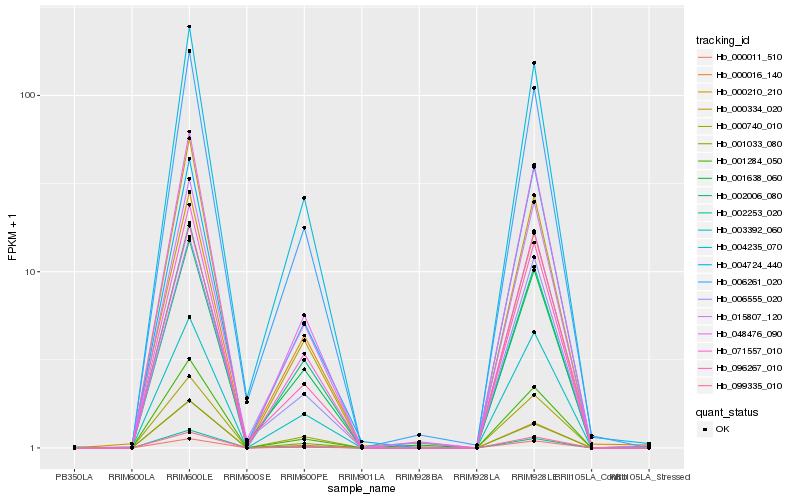
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_099335_010 |
0.0245508984 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 3 |
Hb_000740_010 |
0.0336508323 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_071557_010 |
0.0376965916 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 5 |
Hb_000011_510 |
0.0399701571 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 6 |
Hb_001284_050 |
0.0438913828 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 7 |
Hb_006261_020 |
0.0472651295 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 8 |
Hb_004235_070 |
0.0514105094 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 9 |
Hb_003392_060 |
0.0589760185 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 10 |
Hb_015807_120 |
0.0613246103 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_096267_010 |
0.0615407517 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 12 |
Hb_002006_080 |
0.0646504323 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 13 |
Hb_006555_020 |
0.0655780246 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 14 |
Hb_000334_020 |
0.0683168789 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_001638_060 |
0.0687064538 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000210_210 |
0.0739387003 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004724_440 |
0.0784883105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002253_020 |
0.0795891835 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 19 |
Hb_001033_080 |
0.080567067 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 20 |
Hb_000016_140 |
0.0806093686 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |