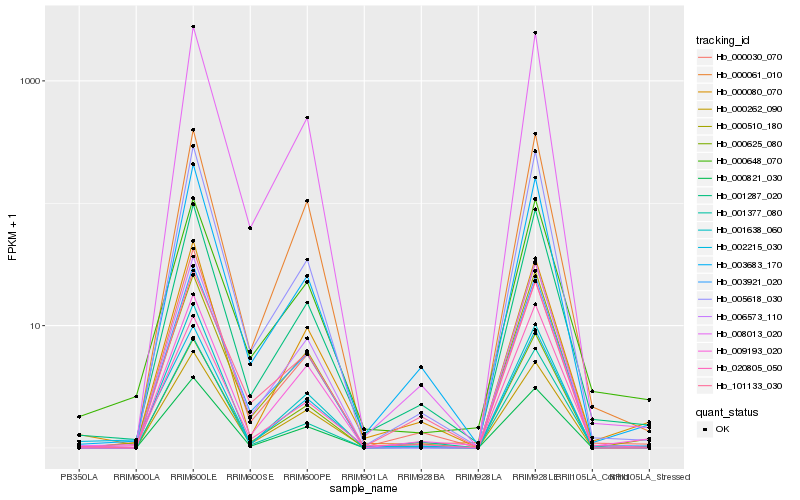
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001287_020 |
0.0 |
- |
- |
SulA [Manihot esculenta] |
| 2 |
Hb_003683_170 |
0.0580442024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000030_070 |
0.0620345238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005618_030 |
0.0727588284 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 5 |
Hb_000080_070 |
0.0785195389 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001377_080 |
0.0820257222 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 7 |
Hb_000510_180 |
0.0847376454 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_000648_070 |
0.08534614 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 9 |
Hb_008013_020 |
0.0855390103 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 10 |
Hb_020805_050 |
0.086121553 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 11 |
Hb_000625_080 |
0.0865971167 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002215_030 |
0.0886970355 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003921_020 |
0.0894968594 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 14 |
Hb_000821_030 |
0.0918930261 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 15 |
Hb_000262_090 |
0.0943814259 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 16 |
Hb_006573_110 |
0.0961586055 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 17 |
Hb_009193_020 |
0.0967552399 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 18 |
Hb_101133_030 |
0.0972271331 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_001638_060 |
0.0993635539 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000061_010 |
0.1001816024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |