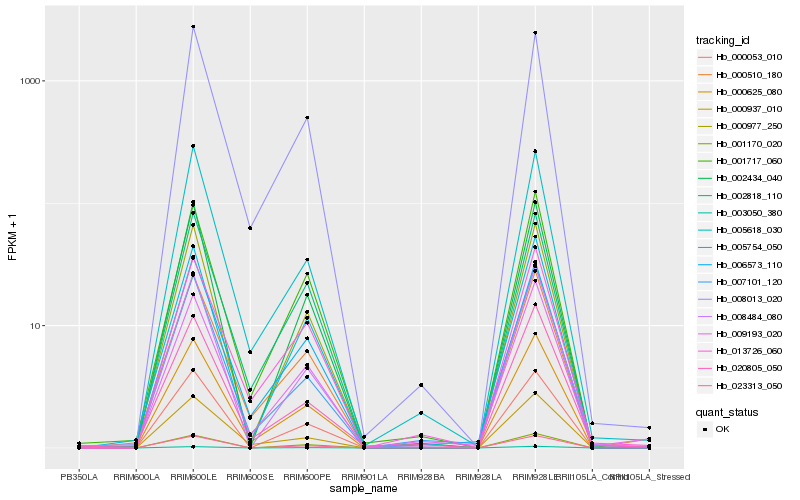
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000625_080 |
0.0 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000510_180 |
0.0449386838 |
- |
- |
unknown [Lotus japonicus] |
| 3 |
Hb_009193_020 |
0.0538133607 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 4 |
Hb_008013_020 |
0.0590553451 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001717_060 |
0.0623813084 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 6 |
Hb_000977_250 |
0.0624480941 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_006573_110 |
0.0631941765 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 8 |
Hb_005618_030 |
0.0655722349 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 9 |
Hb_000937_010 |
0.0658706997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002818_110 |
0.0692203885 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 11 |
Hb_013726_060 |
0.0704269296 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 12 |
Hb_023313_050 |
0.0733353659 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 13 |
Hb_005754_050 |
0.073375342 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 14 |
Hb_002434_040 |
0.0746293121 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 15 |
Hb_000053_010 |
0.0759411001 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 16 |
Hb_001170_020 |
0.0761396342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008484_080 |
0.0765064195 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_007101_120 |
0.0768672723 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 19 |
Hb_003050_380 |
0.0778346361 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 20 |
Hb_020805_050 |
0.0782928329 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |