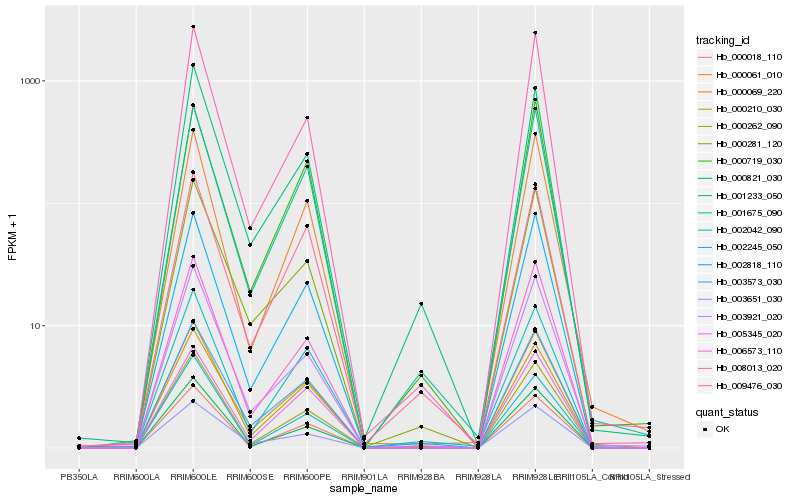
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_030 |
0.0 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 2 |
Hb_000262_090 |
0.0391766025 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 3 |
Hb_003573_030 |
0.0410126865 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 4 |
Hb_002818_110 |
0.0420089168 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 5 |
Hb_001675_090 |
0.0435672518 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000821_030 |
0.0497509646 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 7 |
Hb_006573_110 |
0.0501649839 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 8 |
Hb_008013_020 |
0.0513893237 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000018_110 |
0.052763411 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 10 |
Hb_003651_030 |
0.0543339268 |
- |
- |
lyase, putative [Ricinus communis] |
| 11 |
Hb_000061_010 |
0.0550770392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000281_120 |
0.0555599702 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_002042_090 |
0.0576284268 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 14 |
Hb_005345_020 |
0.0584467993 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 15 |
Hb_000069_220 |
0.0612919227 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 16 |
Hb_001233_050 |
0.0643251931 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003921_020 |
0.0645676566 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 18 |
Hb_009476_030 |
0.0687817825 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 19 |
Hb_002245_050 |
0.0692652645 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000719_030 |
0.0699376464 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |