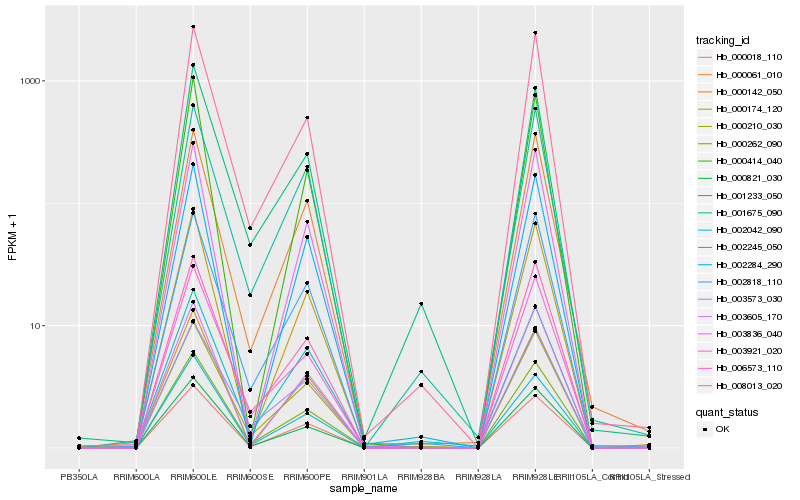
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000262_090 |
0.0 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 2 |
Hb_000821_030 |
0.0328091069 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 3 |
Hb_000210_030 |
0.0391766025 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 4 |
Hb_008013_020 |
0.0432614816 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006573_110 |
0.0456619188 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 6 |
Hb_000018_110 |
0.0526659479 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 7 |
Hb_002818_110 |
0.05474744 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 8 |
Hb_002245_050 |
0.0563663394 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000061_010 |
0.0568088289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000174_120 |
0.0574212774 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_003921_020 |
0.0603703344 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 12 |
Hb_003605_170 |
0.0649378592 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 13 |
Hb_000142_050 |
0.066384223 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 14 |
Hb_003836_040 |
0.0678393038 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 15 |
Hb_002284_290 |
0.0680428804 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 16 |
Hb_003573_030 |
0.0696975928 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 17 |
Hb_001233_050 |
0.0704869665 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001675_090 |
0.0705445276 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000414_040 |
0.0710213637 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 20 |
Hb_002042_090 |
0.071518466 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |