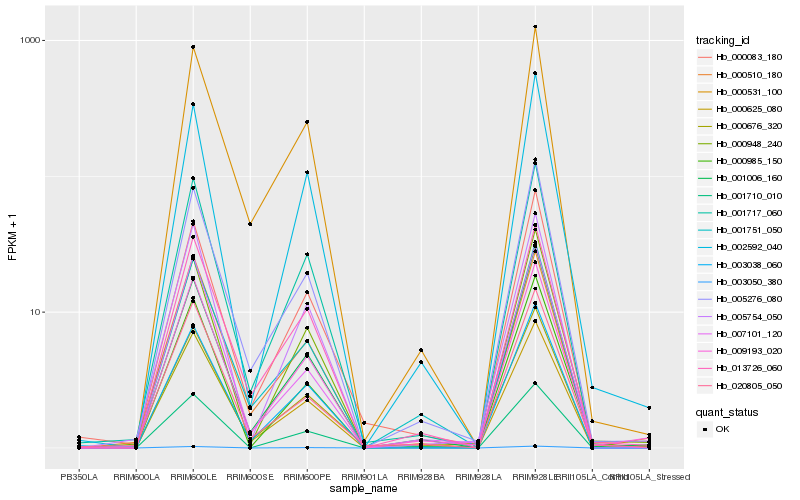
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_020 |
0.0 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 2 |
Hb_000625_080 |
0.0538133607 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005276_080 |
0.0635132001 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000676_320 |
0.0662617023 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 5 |
Hb_013726_060 |
0.0679587025 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 6 |
Hb_000510_180 |
0.0682336685 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_001717_060 |
0.0691606827 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 8 |
Hb_001006_160 |
0.073452797 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000531_100 |
0.0749660159 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_005754_050 |
0.0777582189 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 11 |
Hb_002592_040 |
0.0783501642 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 12 |
Hb_001710_010 |
0.0811357105 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 13 |
Hb_003038_060 |
0.0818827605 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 14 |
Hb_007101_120 |
0.0823710339 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 15 |
Hb_000083_180 |
0.0833597601 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 16 |
Hb_001751_050 |
0.0834959841 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000985_150 |
0.0839548342 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 18 |
Hb_000948_240 |
0.0844682042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_020805_050 |
0.0854373143 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 20 |
Hb_003050_380 |
0.0881688899 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |