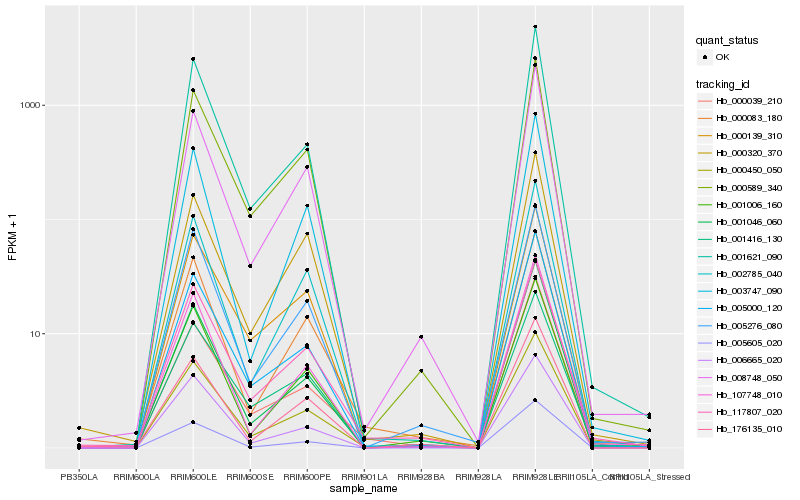
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000450_050 |
0.0 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 2 |
Hb_000589_340 |
0.0440520975 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_001621_090 |
0.0490558861 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 4 |
Hb_005000_120 |
0.0582607916 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 5 |
Hb_005276_080 |
0.058748385 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006665_020 |
0.059909384 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 7 |
Hb_008748_050 |
0.0612298654 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_002785_040 |
0.0616696865 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 9 |
Hb_117807_020 |
0.0652415025 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001006_160 |
0.065308711 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_001416_130 |
0.0672614559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000139_310 |
0.0732770138 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005605_020 |
0.0735345959 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 14 |
Hb_000083_180 |
0.0735870032 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 15 |
Hb_000039_210 |
0.074276485 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 16 |
Hb_003747_090 |
0.0753090805 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001046_060 |
0.0766292839 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_176135_010 |
0.0772912782 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 19 |
Hb_107748_010 |
0.0783071636 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000320_370 |
0.0784832749 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |