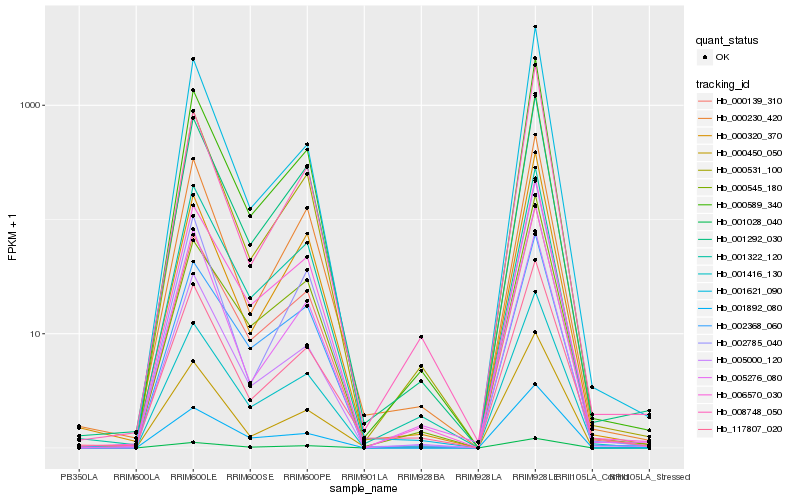
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_340 |
0.0 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 2 |
Hb_000139_310 |
0.0361078419 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001416_130 |
0.0376090158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000450_050 |
0.0440520975 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 5 |
Hb_006570_030 |
0.0535613686 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005276_080 |
0.0618022899 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002785_040 |
0.0624228982 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 8 |
Hb_001028_040 |
0.0632425377 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 9 |
Hb_001292_030 |
0.0635332232 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000531_100 |
0.0636219389 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000320_370 |
0.0646822413 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 12 |
Hb_117807_020 |
0.0654943883 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000230_420 |
0.0658257666 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 14 |
Hb_001621_090 |
0.0665654738 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 15 |
Hb_001892_080 |
0.0688133207 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 16 |
Hb_001322_120 |
0.0703440981 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 17 |
Hb_000545_180 |
0.0704529984 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_005000_120 |
0.0709345799 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 19 |
Hb_008748_050 |
0.0710611219 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 20 |
Hb_002368_060 |
0.0734604646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |