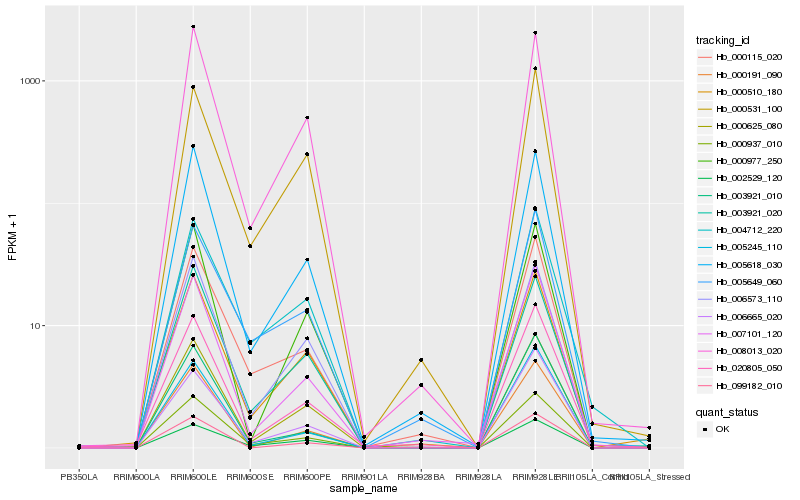
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000937_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000115_020 |
0.0557521792 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_005618_030 |
0.0632141292 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 4 |
Hb_000625_080 |
0.0658706997 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008013_020 |
0.0713970871 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006573_110 |
0.072726375 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 7 |
Hb_007101_120 |
0.0762009031 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 8 |
Hb_000510_180 |
0.0782780412 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_003921_020 |
0.0792498094 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 10 |
Hb_000191_090 |
0.0799422971 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 11 |
Hb_020805_050 |
0.0830584034 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 12 |
Hb_003921_010 |
0.0863174371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005649_060 |
0.0880468901 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 14 |
Hb_002529_120 |
0.0886377182 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 15 |
Hb_005245_110 |
0.0888144085 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 16 |
Hb_004712_220 |
0.0901805245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006665_020 |
0.0906063968 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_000531_100 |
0.0927939648 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_099182_010 |
0.0932505481 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 20 |
Hb_000977_250 |
0.0937462662 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |