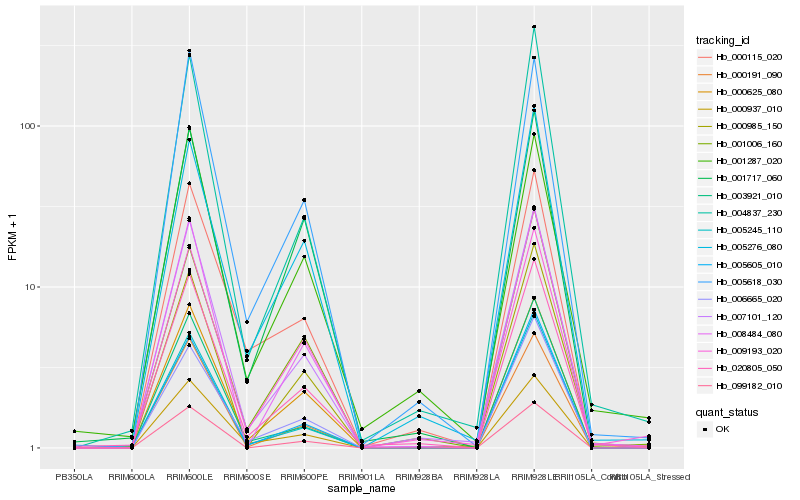
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020805_050 |
0.0 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 2 |
Hb_007101_120 |
0.0565661136 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 3 |
Hb_004837_230 |
0.0660723687 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_008484_080 |
0.0771833893 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000625_080 |
0.0782928329 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006665_020 |
0.0813430371 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 7 |
Hb_000191_090 |
0.0815935041 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 8 |
Hb_005245_110 |
0.0822153141 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 9 |
Hb_005618_030 |
0.0829086096 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 10 |
Hb_000937_010 |
0.0830584034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000985_150 |
0.0853942626 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 12 |
Hb_009193_020 |
0.0854373143 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 13 |
Hb_005276_080 |
0.085830743 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001287_020 |
0.086121553 |
- |
- |
SulA [Manihot esculenta] |
| 15 |
Hb_005605_010 |
0.0874366548 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 16 |
Hb_003921_010 |
0.0888680125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_099182_010 |
0.0900478419 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 18 |
Hb_000115_020 |
0.0911427785 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 19 |
Hb_001006_160 |
0.0923950158 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_001717_060 |
0.09258789 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |