| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
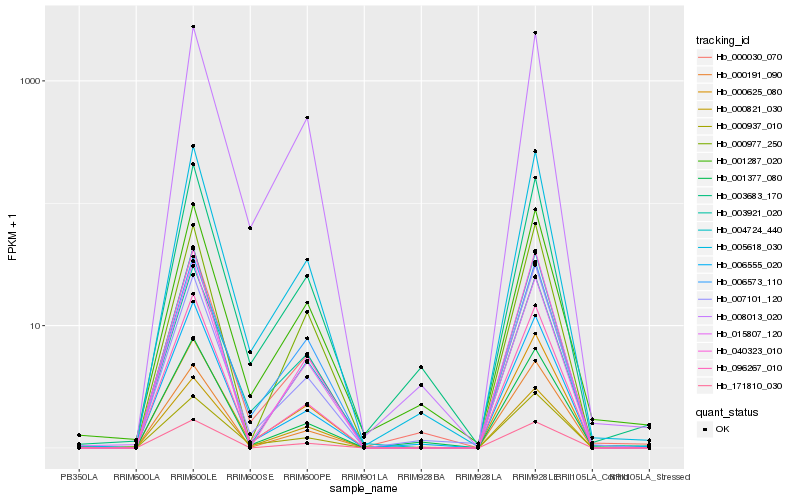
Hb_005618_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 2 |
Hb_000030_070 |
0.0484210455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008013_020 |
0.0542353408 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003683_170 |
0.0617781677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006555_020 |
0.0625253409 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 6 |
Hb_096267_010 |
0.0629947959 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 7 |
Hb_000937_010 |
0.0632141292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003921_020 |
0.0648309859 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 9 |
Hb_000625_080 |
0.0655722349 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006573_110 |
0.0668496803 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 11 |
Hb_004724_440 |
0.0682435858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000821_030 |
0.0709309018 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 13 |
Hb_015807_120 |
0.0709543391 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001377_080 |
0.0715689322 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 15 |
Hb_001287_020 |
0.0727588284 |
- |
- |
SulA [Manihot esculenta] |
| 16 |
Hb_000191_090 |
0.0729686244 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 17 |
Hb_007101_120 |
0.073224418 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 18 |
Hb_171810_030 |
0.0747650404 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 19 |
Hb_040323_010 |
0.0747852368 |
- |
- |
- |
| 20 |
Hb_000977_250 |
0.0788651982 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |