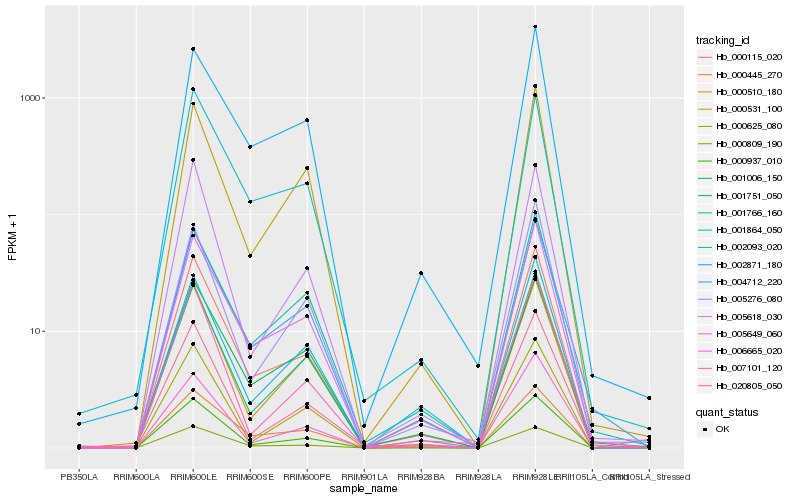
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_020 |
0.0 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 2 |
Hb_005649_060 |
0.053568076 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 3 |
Hb_000937_010 |
0.0557521792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000809_190 |
0.0797034908 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 5 |
Hb_000445_270 |
0.0817868786 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004712_220 |
0.0822765492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001751_050 |
0.0832807421 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001006_150 |
0.0859826093 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_006665_020 |
0.0876259058 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 10 |
Hb_002871_180 |
0.0890714588 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001766_160 |
0.0909961632 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 12 |
Hb_020805_050 |
0.0911427785 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 13 |
Hb_007101_120 |
0.0913911983 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 14 |
Hb_001864_050 |
0.0915485217 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 15 |
Hb_005618_030 |
0.091573899 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 16 |
Hb_000531_100 |
0.0917914073 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000625_080 |
0.0925820598 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000510_180 |
0.0936204182 |
- |
- |
unknown [Lotus japonicus] |
| 19 |
Hb_002093_020 |
0.0936812837 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 20 |
Hb_005276_080 |
0.094563223 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |